SMS Photo Physics Herten - Events
advertisement
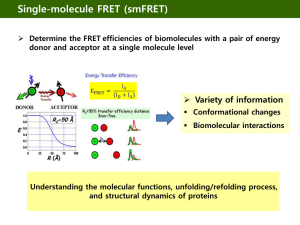
Single molecules & Photo-physics Dirk-Peter Herten Heidelberg University EMBO Course: F-Techniques Heidelberg, 23. -27.9.2009 Molecules Models Human models Modeling Individual Individual Individual Model Average deduce Model Ensemble = Average Why single molecules? • Resolve molecular heterogeneities – Static heterogeneties (Subpopulations) – Dynamic heterogeneities (e.g. transitions between different conformers) – Resolve rare / hidden events – Measure kinetics in thermodynamic equilibrium • Ultimate limit of analytical sensitivity Single-molecule techniques Mechanical selection: Near-field techniques: NSOM, AFM (Surfaces) Spectral selection Solid-state techniques (Glases) Dilute & Select Spatial selection: Far-field techniques: SMFS, Ftechniques … Spatial selection Confocal fluorescence microscopy: Total-internal reflection fluorescence microscopy (TIRFM): Diffraction limited excitation/detection (~ 1fl) Evanescent wave (~ 100 nm) Laser Detektor (APD) Laser CCD Reject background What does a single molecule look like? This is a single molecule! Enzymatic catalysis KM E+S ES kcat E+P • Substrate binding association • Conformational change • Allosteric interaction • Co-enzyme binding • Catalytic conversion ‘Chemical reaction’ • Series of elementary step (protonation, cleavage, deprotonation, substitution, oxidation, ….) • Product dissociation • … Molecular transitions Single-molecule fluorescence spectroscopy Location Conformation e- Constitution Redox-state + Objective: Connect molecular states to changes in fluorescence emission. Photo-physics / Photochemistry Photo-physics & Photo-chemistry • Fluorescence Resonance Energy Transfer (FRET) • Photo-induced Electron Transfer (PET) • Redox Reactions (Oxidation / Reduction) • Protonation • Charge-Transfer Bands • …. Photo-physics & Photo-chemistry • Fluorescence Resonance Energy Transfer (FRET) • Photo-induced Electron Transfer (PET) • Redox Reactions (Oxidation / Reduction) • Protonation • Charge-Transfer Bands • …. Redox-state Lu et al. Science 282 (1998), 1877-1882 Photo-physics & Photo-chemistry • Fluorescence Resonance Energy Transfer (FRET) • Photo-induced Electron Transfer (PET) • Redox Reactions (Oxidation / Reduction) • Protonation • Charge-Transfer Bands • …. Photo-induced electron transfer (PET) OH O H3C + N O N N Dye Energy LUMO HOMO Reducing reagent 1. Excitation 2. Reduction (ET1) 3. Recombination (ET2) short range effect (contact pair) Folding the Tryptophan Cage Neuweiler et al., Angew. Chem. 2003 Photo-physics & Photo-chemistry • Fluorescence Resonance Energy Transfer (FRET) • Photo-induced Electron Transfer (PET) • Redox Reactions (Oxidation / Reduction) • Protonation • Charge-Transfer Bands • …. Fluorescence resonance energy transfer (FRET) • Non-radiative energy transfer from an excited donor to an acceptor dye. • Strong distance dependence on the range of 2 – 8 nm. FRET – Distance FRET – Orientation κ2 – Orientational parameter FRET – Spectral Overlap D Similar energy levels A Single pair FRET E FRET I – intensity I* – background corrected intensity γ – crosstalk correction IA intensity I *A = * I A + gI D* • Solution (confocal microscope): • Limited by diffusion (1 – 2 ms) ID time countrate / kHz 80 60 40 20 0 0 5 10 time / s 15 20 • Immobilization: • time-resolved studies can resolve (dynamic) heterogeneities and kinetics. • limited by photo-bleaching. Zero FRET efficiency A Alternating Laser Excitation (ALEX) Example: F1F0-ATPase 3 • Site-specific mutagenesis & labeling. • Control of functionality. • Reconstitution in vesicel membranes slow down diffusion extended observation time Dietz et al., Nature Meth. Struct. Biol. 11 (2004), 135 Directionality and kinetics of F1F0-ATPase rotation Hydrolysis of ATP: Synthesis of ATP: High – Medium – Low Low – Medium - High 11-06-02 b64bisCy5-g-TMR @ ATP synthesis 1,0 1,0 1,0 0,8 0,8 0,8 0,8 0,6 0,6 0,6 0,6 0,4 0,4 0,4 0,4 0,2 0,2 0,2 0,2 0,0 0,0 0,0 60 0,0 60 40 40 50 50 40 40 30 30 20 20 10 10 30 30 20 20 10 10 0 0 5200 5300 5400 5500 5600 5700 5800 5900 6000 time / ms proximity factor 1,0 photon counts per ms photon counts per ms proximity factor 10-06-02 b64bisCy5-g-TMR @ ATP hydrolysis 0 700 800 900 0 1000 1100 1200 1300 1400 1500 time / ms Photo-physics is key to SMFS • Förster Resonance Energy Transfer (FRET) distance dependence: 2 – 8 nm Location Conformation - e+ eConstitution • Photo-induced Electron Transfer (PET) distance dependence: < 1nm Redox-state +. • Charge-transfer (MO interaction): direct / transfer • Changes in the chromophore: direct •… Combining photo-physical processes ATTO 520 Double stranded DNA: Stiff (persistence length of ~ 50 nm) Defined distances (molecular ruler) Cy5 Established labeling procedures Ideal scaffold to test photo-physical reactions Kumbakhar et al., ChemPhysChem 2009 Balancing FRET and ET FRET/ET 1 EET ET FRET 2 3 4 5 6 7 Φr 0.15 0.19 0.14 0.17 0.19 0.34 0.46 1.00 ED 0.80 0.51 0.10 - 0.91 0.73 0.51 - EA 0.48 0.18 0.03 - 0.39 0.43 0.34 - Bulk data suggests competition between ET and FRET. Proximity / EFRET 8 spFRET experiments FRET Donor-only 40 B 30 A Count Rate, kHz 20 10 0 0 3 6 9 12 15 18 21 30 24 C 20 10 0 0 3 6 9 12 15 18 Time (second) E FRET I *A = * I A + gI D* Acceptor bleaching Donor bleaching FRET efficiency distributions Normalised number of occurence 5 1 6 2 7 3 0.4 0.6 0.8 1.0 FRET Efficiency (E) Ensemble: 2 populations, (FRET & ET); Single-molecule: 1 population (FRET) Fluorescence fluctuations FRET-only 40 B 30 Count Rate, kHz 20 10 0 0 9 6 3 12 15 21 18 30 24 C 20 10 0 0 3 6 9 12 Time (second) FRET-ET 15 18 Fluorescence fluctuations: - After acceptor bleaching - Only in presence of guanine Fluctuation kinetics A <on> / ms (2') <off> / ms B 6 3 (3') 2 (3) X (2) (1) 21 (1) 18 15 12 (2) 9 (2') (3') (3) 6 3.5 4.0 4.5 R / nm 5.0 5.5 1, 2, 3 2’, 3’ (mismatches) DNA breathing The longer the p-stack the more probable ET interrupted by breathing or partial unzipping or by charge trapping. Summary • Photo-physics is key – FRET: Distance, Spectral Overlap, Orientation – PET: Short distance effect – Redox Reaction Similar Energies / Redox potentials … • Combining PET & FRET in dsDNA: – SMFS reveals molecular heterogeneity – fluorescence fluctuations indicate breathing of dsDNA and electron transfer through π-stack BARC, Mumbai, India Haridas Pal Manoj Kumbhakar Alex Kiel Kostas Lymperopoulos Daniel Siegberg Haisen Ta Tanja Erhard Daniel Barzan Christina Spassova Jessica Balbo Michael Schwering Anne Seefeld Anton Kurz Arina Rybina Thank you! EXC 81