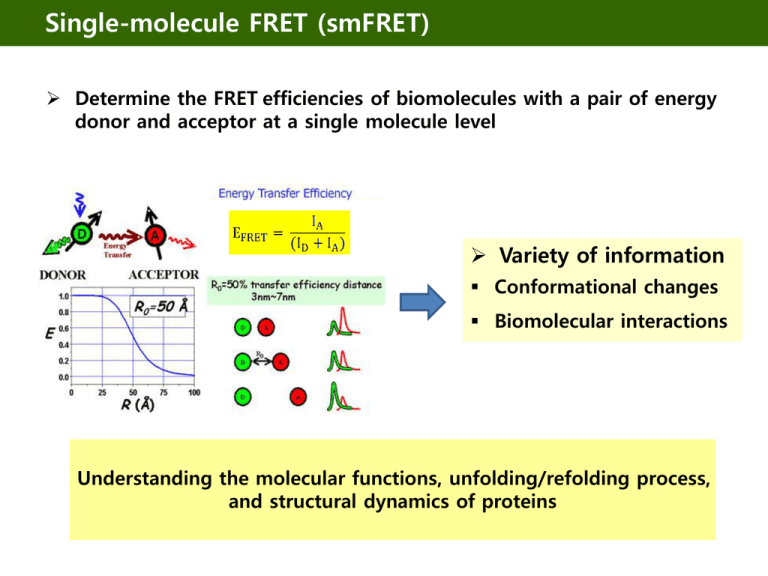
Single-molecule FRET (smFRET)
Determine the FRET efficiencies of biomolecules with a pair of energy
donor and acceptor at a single molecule level
Variety of information
Conformational changes
Biomolecular interactions
Understanding the molecular functions, unfolding/refolding process,
and structural dynamics of proteins
Major issue in biosciences
Ensemble average
Single molecule-based technologies enabling us to
manipulate and probe individual molecules
Answer many of fundamental biological questions :
- Protein functions : Dynamics and recognition
- Biomolecular interactions
- Biological phenomenon
Single molecule FRET
Replication
Recombination
Transcription
Translation
RNA folding and catalysis
Protein folding and
conformational change
• Motor proteins
• Signal transduction
•
•
•
•
•
•
Measure the extent of non-radiative energy transfer between
the two fluorescent dye molecules, donor and acceptor
Intervening distance which can be estimated from the ratio
of acceptor intensity to total emission intensity
ex) Conformational dynamics of single molecules in real time
by tracking FRET changes
Advantages of FRET technique
- A ratiometric method that allows measurement of the internal distance in
the molecular frame with minimized instrumental noise and drift
- Powerful in revealing population distribution of inter-dye distance
FRET- based single molecule analysis
Experimental design
Imaging surface immobilized molecules with the aid of total
internal reflection (TIR) microscopy enabling high throughput
data sampling
Single-molecule fluorescence dye
- Bright ( Extinction coeff. > 50,000 /M/cm; quantum yield > 0.1)
- Photostable with minimal photophysical or chemical and
aggregation effects
- Small and water soluble with sufficient forms of bio-conjugation
chemistries
smFRET pair
Large spectral separation between donor and acceptor emissions
Similar quantum yields and detection efficiencies
cf) Fluorescent proteins : low stability, photoinduced blinking
Quantum dots : large size (>20 nm), lack of a monovalent
conjugation scheme
The most popular single-molecule fluorephores : small (< 1nm)
organic dye
Enhancing photostability
Molecular oxygen : effective quencher of a dye’s unfavorable triplet
state, but a source of a highly reactive species that ultimately causes
photo-bleaching
Vitamin E analogue, Trolox, : excellent triplet-state quencher,
suppressing blinking and stimulating long-lasting emission of the
popular cyanine dyes
The most popular enzymatic oxygen scavenging system:
a mix of glucose oxidase (165 U/mL), catalase(2,170 U/ml),
b-D-glucose (0.4 % w/w)
Conjugation
Schematics for single-molecule FRET analysis
Prism-type Total Internal Reflection Fluorescence (TIRF) microscope
ligand
Detection of fluorescence intensities from two dyes
Electron-multiplying charge-coupled device(EM-CCD) cameras
Usual setup : high quantum efficiency(85-95%) in the 450-700nm range, low
effective readout noise (<1 electron r.m.s.) even at the fastest readout
speed (> 10 MHz), fast vertical shift speed((< 1 us/row)
To achieve adequate signal-to-noise ratio, ~ 100 total photons need to be
detected. More than 105 photons can be collected from single dye
molecules before photobleaching, more than 103 data points can be
obtained.
FRET efficiency : IA/(IA + ID),
IA = acceptor intensity, ID = donor intensity
- Provide only an approximate indicator of the inter-dye distance because
of uncertainty in the orientation factor between the two fluorophores
and the required instrumental corrections
- Correction factor : difference in quantum yield and detection efficiency
between donor and acceptor
Immobilization of dye-labeled biomolecules on a surface
Sample chamber
Limitations of sm FRET
Attachment of at least two intrinsic dyes to the molecule of
interest
Weakly interacting fluorescent species are difficult to study
Insensitive to distance change outside the 2 ~8 nm inter-dye
distance
Time resolution is limited by the frame rate of the CCD camera
( in best case = 1 ms)
Absolute distance estimation is challenging because of the
dependence of the fluorescence properies and energy transfer
on the environment and orientation of the dyes
Intrinsic motions along an enzymatic reaction trajectory
Adnylate kinases : enzymes that maintain the cellular equilibrium
concentration of adenylate nucleotides by catalyzing the
reversible conversion of ATP and AMP into two ADP molecules
•
Composed of a core domain plus ATP and AMP lids
Henzler-Wildman et al., Nature, 450, 838-850 (2007)
Challenging issue in smFRET
Labeling of proteins with two fluorescent dyes (donor and acceptor)
Most common conventional method for labeling involves:
- Introduction of two cysteine residues into desired sites on proteins
Dye heterogeneity
Limited to the nucleic acid-interacting proteins and a subset of
proteins that are tolerable to cysteine mutations
Site-specific dual-labeling of proteins
Genetic code expansion
Incorporation of unnatural amino acids:
- Broadening the chemical and biological functionalities
- Proteins containing UAAs have novel property
Nonsense codon suppression method
Introduce a stop codon (TAG) at a specific site of a target gene
Bioorthogonal aminoacyl tRNA synthetase and tRNA pair for UAA
Expression of protein containing UAA
tRNA synthetase
Met
Ribosome
Arg
AGC
TAG
tRNA
His
Ser
UAA
Site-directed
mutagenesis
Transcription
mRNA
Nonsense codon
Translation
Site-specific labeling using unnatural amino acid
Dual-labeling of maltose binding protein (MBP)
Incorporation of azido-phenylalanine into Lys42 via an amber codon (TAG)
- Engineered tyrosyl-tRNA synthetase/tRNACUA of Methanococcus jannaschi
- Conjugation with Cy5-alkyne by click chemistry
Incorporation of cysteine residue into Lys370
wt
Lys42AzF/
Lys42AzF Lys370Cys
Seo et al., Anal Chem (2011)
Single-molecule FRET measurements
Prism-type Total Internal Reflection Fluorescence (TIRF) microscope
ligand
Time resolution: 50 ms
smFRET analysis of dual-labeled MBP
Dual-labeled MBP using UAA
Histograms of FRET efficiency
Much clearer picture for the folded and unfolded states in smFRET
Seo et al., Anal Chem (2011)
Site-specific dual-labeling using two UAAs for smFRET analysis
Incorporation of p-acetylphenylalanine and alkynelysine into Thr34 and Gly113 on Calmodulin
- Evolution of Methanosarcina mazei pyrrolysyl-tRNA synthetase (PylRS) for improved
incorporation of AlK : L301M and Y306L
- p-Acetylphenylalanyl-tRNA synthetase/tRNACUA
Conjugation of two dyes (Cy3-hydrazide and Cy5-azide) via ketone-oxyamine and click reactions
ρ-acetylphenylalanine (AcF)
Fluorescence scan
Calmodulin
Lane 1 : CaM
Lane 2 : Dual-labeled CaM
Alkynelysine (AlK)
Analysis of conformational change by smFRET
M13
Ca2+
Histograms of FRET efficiency for M13-induced conformational change of CaM