Biochem-5012.2B - Center for Structural Biology
advertisement
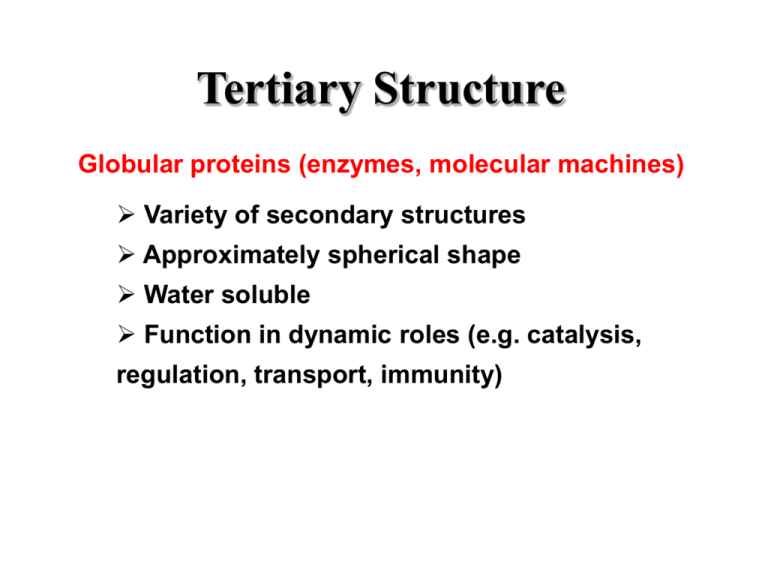
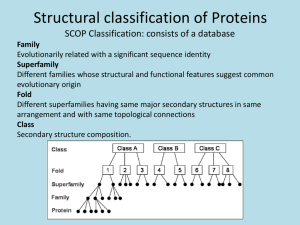
Tertiary Structure Globular proteins (enzymes, molecular machines) Variety of secondary structures Approximately spherical shape Water soluble Function in dynamic roles (e.g. catalysis, regulation, transport, immunity) Tertiary Structure Fibrous Proteins (fibrils, structural proteins) One dominating secondary structure Typically narrow, rod-like shape Poor water solubility Function in structural roles (e.g. cytoskeleton, bone, skin) Tertiary Structure Membrane Proteins (receptors, channels) Inserted into (through) membranes Multi-domain- membrane spanning, cytoplasmic, and extra-cellular domains Poor water solubility Function in cell communication (e.g. cell signaling) Quaternary Structure Definition: Organization of multiple chain associations Oligomerization- Homo (self), Hetero (different) Used in organizing single proteins and protein machines Specific structures result from long-range interactions Electrostatic (charged) interactions Hydrogen bonds (OH, N H, S H) Hydrophobic interactions Disulfides only VERY infrequently Quaternary Structure The classic example- hemoglobin a2-b2 Protein Folding Folded proteins are only marginally stable!! ~0.4 kJ•mol-1 required to unfold (cf. ~20/H-bond) Balance of loss of entropy and stabilizing forces Protein fold is specified by sequence Reversible reaction- denature (fold)/renature Even single mutations can cause changes Recent discovery that amyloid diseases (eg. CJD, Alzheimer) are due to unstable protein folding Protein Folding The hydrophobic effect is the major driving force Hydrophobic side chains cluster/exclude water Release of water cages in unfolded state Other Forces stabilizing protein structure Hydrogen bonds Electrostatic interactions Chemical cross links- Disulfides, metal ions Protein Folding Random folding has too many possibilities Backbone restricted but side chains not A 100 residue protein would require 1087 s to search all conformations (age of universe < 1018 s) Most proteins fold in less than 10 s!! *Proteins fold along specific pathways* Protein Folding Pathways Usual order of folding events Secondary structures formed quickly (local) Secondary structures aggregate to form motifs Hydrophobic collapse to form domains Coalescence of domains Molecular chaperones assist folding in-vivo Complexity of large chains/multi-domains Cellular environment is rich in interacting molecules Chaperones sequester proteins and allow time to fold Relationships Among Proteins I. Homologous: very similar sequence (cytochrome c) Same structure Same function Modeling structure from homology II. Similar function- different sequence (dehydrogenases) One domain same structure One domain different III. Similar structure- different function (cf. thioredoxin) Same 3-D structure Not same function Relationships Among Proteins Many sequences can give same structure Side chain pattern more important than sequence When homology is high (>50%), likely to have same structure and function (structural genomics) Cores conserved Surfaces and loops more variable *3-D shape more conserved than sequence* *There are a limited number of structural frameworks*