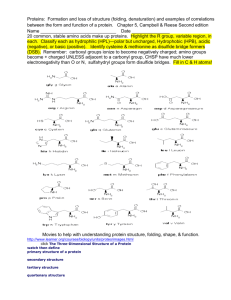
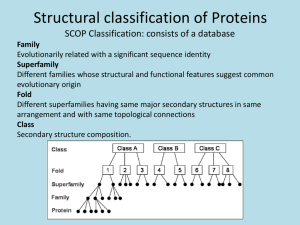
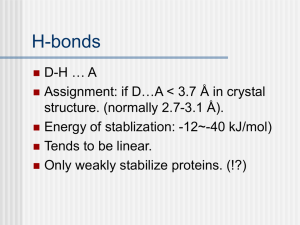
Relationship between amino acids sequences and protein structures
advertisement

Alexander Kister University of Medicine and Dentistry of New Jersey, SHRP, Department of Health Informatics, Newark, NJ, 07107, USA Relationship between amino acids sequences and protein structures: folding patterns and sequence patterns. To describe the main structural regularities - folding pattern we introduced a supersecondary structure (SSS) unit of beta proteins called the ‘‘strandon’’. Analysis of sandwich-like proteins revealed two specific folding patterns, which describe the arrangement of strandons and strands in the protein domains. It enables us to construct a novel structural classification scheme in the recently created SSS Protein database (http://binfs.umdnj.edu/sssdb/). The second goal of this study was to evaluate the hypothesis that proteins from different families and with very low sequence similarities but with an identical SSS have a common sequence pattern. To find common sequence regularities a new algorithm of the multiple sequences alignment was developed. The proposed method involves ‘projecting’ common supersecondary structural characteristics onto the sequence to reveal key residues. The results of the alignment revealed conserved positions. It was shown that residues at these positions form an SSS-specific sequence pattern and could be used for protein classification as an amino acid ‘tag’ that brands a sequence as having a particular SSS. The important conclusion follows from the results of this work: the protein sequence/structure relationship cuts both ways. That is both the sequence of amino acids determines the folding pattern and the structure defines key positions in sequences and residues at these positions. Reference: PNAS (2006) vol. 103: 4107-4110; Proteins (2007) vol. 68: 915–921; PNAS (2008) vol. 105: 9233-9237; Protein Sci. (2008) the manuscript is submitted