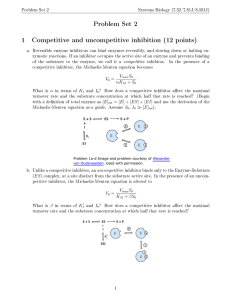
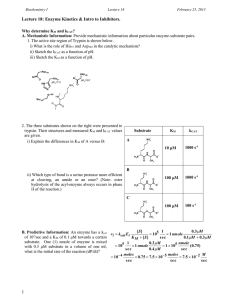
Problem Set 3 - Chemistry at Caltech
advertisement
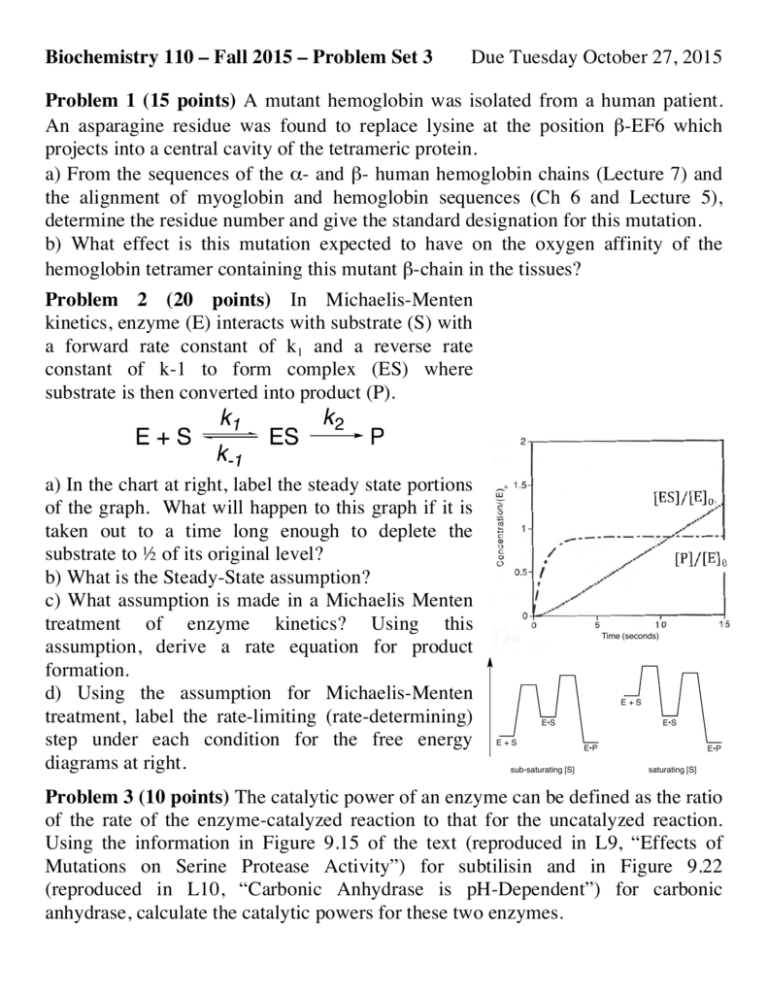
Biochemistry 110 – Fall 2015 – Problem Set 3 Due Tuesday October 27, 2015 Problem 1 (15 points) A mutant hemoglobin was isolated from a human patient. An asparagine residue was found to replace lysine at the position β-EF6 which projects into a central cavity of the tetrameric protein. a) From the sequences of the α- and β- human hemoglobin chains (Lecture 7) and the alignment of myoglobin and hemoglobin sequences (Ch 6 and Lecture 5), determine the residue number and give the standard designation for this mutation. b) What effect is this mutation expected to have on the oxygen affinity of the hemoglobin tetramer containing this mutant β-chain in the tissues? Problem 2 (20 points) In Michaelis-Menten kinetics, enzyme (E) interacts with substrate (S) with a forward rate constant of k1 and a reverse rate constant of k-1 to form complex (ES) where substrate is then converted into product (P). k2 ! a) In the chart at right, label the steady state portions of the graph. What will happen to this graph if it is taken out to a time long enough to deplete the substrate to ½ of its original level? b) What is the Steady-State assumption? c) What assumption is made in a Michaelis Menten treatment of enzyme kinetics? Using this assumption, derive a rate equation for product formation. d) Using the assumption for Michaelis-Menten treatment, label the rate-limiting (rate-determining) step under each condition for the free energy diagrams at right. Time (seconds) E+S E•S E+S sub-saturating [S] E•S E•P E•P saturating [S] Problem 3 (10 points) The catalytic power of an enzyme can be defined as the ratio of the rate of the enzyme-catalyzed reaction to that for the uncatalyzed reaction. Using the information in Figure 9.15 of the text (reproduced in L9, “Effects of Mutations on Serine Protease Activity”) for subtilisin and in Figure 9.22 (reproduced in L10, “Carbonic Anhydrase is pH-Dependent”) for carbonic anhydrase, calculate the catalytic powers for these two enzymes. Problem 4 (15 points) The structure Glu 35 of lysozyme was published in 1965, HO but the mechanism by which O lysozyme catalyzes the replacement O O Lysozyme O-NAG OH HO HO NAG O O NAG O O of an N-acetylglucosaminyl group CH NHAc NHAc O CH3 3 HO COOCOO(NAG-OH) on a sugar molecule with water remained a matter of NAG = N-acetylglucosamine Asp 52 debate for several decades. a) The substrate and product of the reaction catalyzed by lysozyme are shown below, along with the two potential catalytic groups (Glu35 and Asp52) that were identified in the early crystal structures of the enzyme. Why is a concerted (SN2type) substitution not a plausible mechanism for this reaction? b) The two alternative mechanisms that were under debate are (1) a doubledisplacement mechanism involving first a carboxylate group, followed by water and (2) a dissociative (SN1-type) substitution. The former was proposed by David Chilton Phillips (who solved the structure of lysozyme) and the latter is currently favored (Vocadlo et al., 2001). Draw these two mechanisms, taking care to show the stereochemistry of the sugar molecule as well as any charges on the atoms. Problem 5 (10 points) (a) During digestion, all of the pancreatic enzymes are activated at virtually the same time. What is the mechanism of activation, and how is the simultaneity of activation achieved ? (b) Pancreatic trypsin inhibitor contains a lysine at position 15, which forms a salt bridge with a residue in the active site of trypsin. What phenotype might you expect in a patient whose pancreatic trypsin inhibitor contains a K15Q mutation ? Rate with Rate with Problem 6 (20 points) The following kinetic [Substrate] no inhibitor inhibitor data were obtained for an enzyme with and mM (mM/min) (mM/min) 1.42 0.200 0.180 without an inhibitor (I). Generate a 0.80 0.180 0.150 Lineweaver-Burk plot with the data and 0.45 0.160 0.120 calculate Vmax and KM for this enzyme in the 0.20 0.120 0.080 presence and absence of inhibitor. What type 0.14 0.100 0.060 0.08 0.070 0.040 of inhibitor is I ? Problem 7 (10 points) The Lipscomb structure (pdb code: 1RAI) is used in the text to display the features of ATCase c6r6 complex and highlight the nature of the “T” and “R” conformations. Recently, an ATCase mutant was obtained with an A241C mutation in all six catalytic subunits. From the RCSB file for the 1RAI “Biological Assembly”, determine the distances between adjacent 241-Cα atoms in the six catalytic peptide chains. Upon disulfide formation, would you expect this mutant to exist in the T or R state?