Document 15549581
advertisement
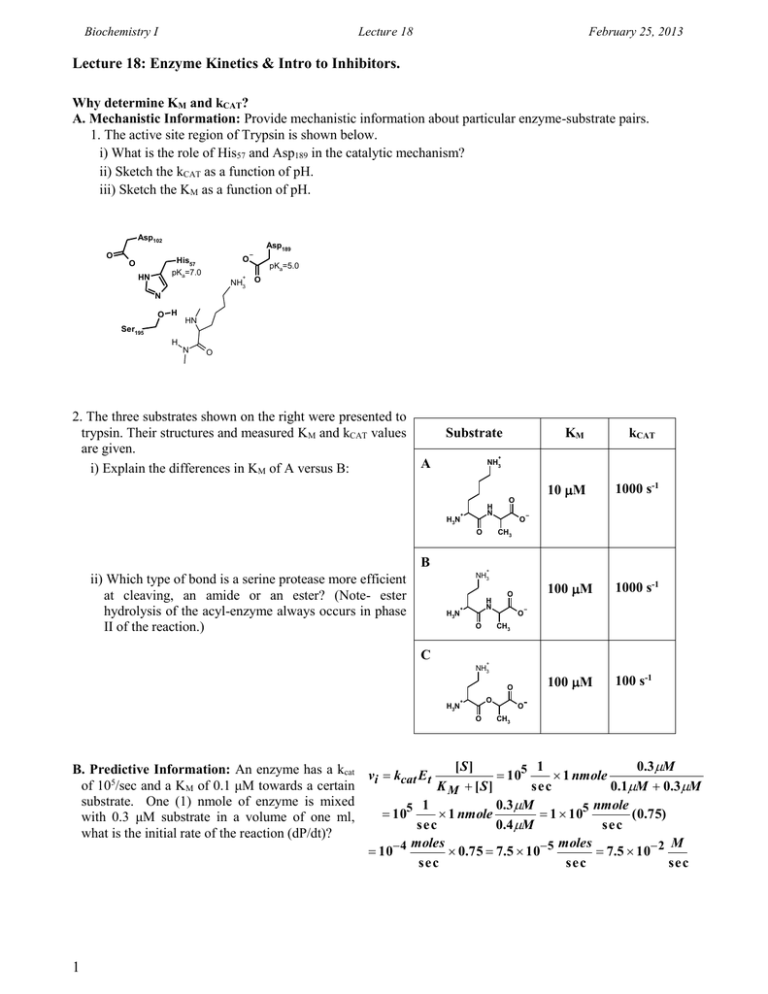
Biochemistry I Lecture 18 February 25, 2013 Lecture 18: Enzyme Kinetics & Intro to Inhibitors. Why determine KM and kCAT? A. Mechanistic Information: Provide mechanistic information about particular enzyme-substrate pairs. 1. The active site region of Trypsin is shown below. i) What is the role of His57 and Asp189 in the catalytic mechanism? ii) Sketch the kCAT as a function of pH. iii) Sketch the KM as a function of pH. Asp102 O Asp189 O His57 pKa=7.0 O HN + NH3 pKa=5.0 O N O H HN Ser195 H N O 2. The three substrates shown on the right were presented to trypsin. Their structures and measured KM and kCAT values are given. i) Explain the differences in KM of A versus B: Substrate kCAT 10 M 1000 s-1 100 M 1000 s-1 100 M 100 s-1 + A NH3 H3N H N + B O O CH3 O + NH3 ii) Which type of bond is a serine protease more efficient at cleaving, an amide or an ester? (Note- ester hydrolysis of the acyl-enzyme always occurs in phase II of the reaction.) KM H3N H N + O CH3 O C O + NH3 O H3N O + O O CH3 [S] 1 0.3M B. Predictive Information: An enzyme has a kcat v k E 105 1 nmole i cat t 5 of 10 /sec and a KM of 0.1 μM towards a certain K M [S] sec 0.1M 0.3M substrate. One (1) nmole of enzyme is mixed 1 0.3M nmole 105 1 nmole 1 105 ( 0.75) with 0.3 μM substrate in a volume of one ml, sec 0.4 M sec what is the initial rate of the reaction (dP/dt)? moles moles M 10 4 0.75 7.5 10 5 7.5 10 2 sec sec sec 1 Biochemistry I Lecture 18 February 25, 2013 Enzyme Inhibitors Studies on Inhibitors are useful for: 1. Mechanistic studies to learn about how enzymes interact with their substrates. 2. Understanding the role of inhibitors in enzyme regulation. 3. Drugs if they inhibit aberrant biochemical reactions: penicillin, ampicillin, etc.:interfere with the synthesis of bacterial cell walls 4. Understanding the role of biological toxins. Amino acid analogs - useful herbicides (i.e. acetylcholine roundup) esterase OH Insecticides - chemicals targeted for insect + O CH3 (Serine esterase) + H3N H3N choline nervous system. Acetylcholine HO O Molecular Mechanism of Inhibitors: A: Suicide Inhibitors Inhibitor binds and forms a covalent bond with the enzyme, inactivating it. Nerve gas sarin modifies the active site serine in the serine esterase acetylcholine esterase. B: Competitive Inhibition 1. Inhibitor binds to the same site on HO O the enzyme as the substrate. H H 2. Inhibitor ONLY binds to the free H H enzyme. 3. Inhibitor usually is structurally HO O very similar to the substrate. For Succinate example, succinate is the normal HO O substrate for the enzyme H H succinate dehydrogenase. Malonate is an effective HO O competitive inhibitor of this enzyme. Malonate 4. The inhibitor can’t undergo the chemical reaction. C: Mixed Inhibition: In this case the inhibitor binds to both [E] and [ES]. The binding site of the inhibitor is not at the active site. However, the inhibitor binding causes a change in the conformation of the protein that affects either substrate binding, the chemical step or both. Allosteric Activators increase substrate binding and/or the rate of the chemical step (kcat). Allosteric inhibitors reduce substrate binding and/or the rate of the chemical step. Both VMAX and KM can be altered by mixed inhibitors since the precise geometry of the active site is altered when the inhibitor is bound. 2 H3C O H3C P O CH3 sarin F O (nerve gas) O H H HO O H3C P O CH3 O Serine HO H3C O Serine CH3 acetic O acid Biochemistry I Lecture 18 February 25, 2013 D. Measuring Inhibitor-Enzyme Affinity: This can be done by measuring the effect of the presence of the inhibitor on the enzyme rate constants, KM and VMAX. Two sets of experiments are performed: 1. Substrate is varied in the absence of the inhibitor, as you would normally do to obtain KM and VMAX. 2. Substrate is varied in the presence of a known, and constant, amount of inhibitor. Competitive Inhibitor: A competitive inhibitor reduces the amount of [E] by the formation of [EI] complex. The inhibitor cannot affect the [ES] complex after it has formed since the inhibitor can no longer bind. E + (ES) S EP (EI) There are two anticipated consequences of this binding mode on the steady-state kinetics: 1. VMAX is unchanged: At high levels of substrate all of the inhibitor can be displaced by substrate, and ET=[ES]. 2. The apparent KM is increased: It requires more substrate to reach 1/2 maximal velocity because some of the enzyme is complexed with inhibitor. A B D C E ES EI [S] < KM A' [S] = KM B' [S] > KM C' [S] >> KM D' 6 5 4 vi 3 2 1 C B C' A B' A' [S] 3 w/o Inh D + Inh D'