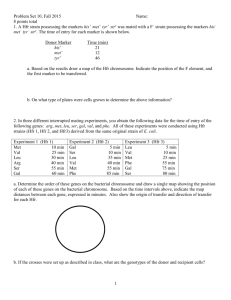
Demonstration
advertisement

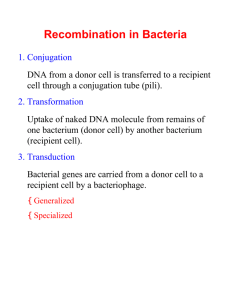
Search for Within This book All books PubMed NCBI » Bookshelf » An Introduction to Genetic Analysis » Gene Transfer in Bacteria and Their Viruses » Bacterial conjugation By agreement with the publisher, this book is accessible by the search feature, but cannot be browsed. Bacterial conjugation 7 Table of Contents In this page This section and subsequent sections describe the discovery of gene transfer in bacteria and explain several types of gene transfer and their use in bacterial genetics. First, we shall consider conjugation, which requires cell-to-cell contact. Conjugation was the first extensively studied method of gene transfer. Discovery of conjugation Requirement for physical contact Discovery of the fertility factor Discovery of conjugation Do bacteria possess any processes similar to sexual reproduction and (F) Hfr strains Determining linkage from interrupted-mating experiments recombination? The question was answered in 1946 by the elegantly simple experimental work of Joshua Lederberg and Edward Tatum, who studied two strains of Escherichia coli with different nutritional requirements. Strain A would grow on a minimal medium only if the medium were supplemented with methionine and biotin; strain B would grow on a minimal medium only if it were supplemented with threonine, leucine, and thiamine. Thus, we can designate Figure 7-2 Demonstration by Lederberg and Tatum of genetic recombination (more...) strain A as met − bio − thr+ leu + thi+ and strain B as met + bio + thr− leu − thi− . Chromosome circularity and integration of F R factors Mechanics of transfer E. coli conjugation cycle Figure 7-2a displays in simplified form the concept of their experiment. Here, Recombination between marker strains A and B are mixed together, and some of the progeny are now wild type, genes after transfer having regained the ability to grow without added nutrients. Figure 7-2b Gradient of transfer illustrates their experiment in more detail. Lederberg and Tatum plated bacteria into dishes containing only unsupplemented minimal medium. Some of the dishes were plated only with strain A bacteria, Determining gene order from gradient of transfer Higher-resolution mapping by some only with strain B bacteria, and some with a mixture of strain A and strain B bacteria that had recombinant frequency in been incubated together for several hours in a liquid medium containing all the supplements. No bacterial crosses colonies arose on plates containing either strain A or strain B alone, showing that back mutations cannot restore prototrophy, the ability to grow on unsupplemented minimal medium. However, the plates that received the mixture of the two strains produced growing colonies at a frequency of 1 in every 10,000,000 cells plated (in scientific notation, 1 × 10−7). This observation suggested that some form of recombination of genes had taken place between the genomes of the two strains to produce prototrophs. Requirement for physical contact It could be suggested that the cells of the two strains do not really exchange genes but instead leak substances that the other cells can absorb and use for growing. This possibility of “cross feeding” was ruled out by Bernard Davis. He constructed a U-tube in which the two arms were separated by a fine filter. The pores of the filter were too small to allow bacteria to pass through but large Figure 7-3 Experiment demonstrating that physical contact between (more...) enough to allow easy passage of the fluid medium and any dissolved substances (Figure 7-3 ). Strain A was put in one arm; strain B in the other. After the strains had been incubated for a while, Davis tested the content of each arm to see if cells had become able to grow on a minimal medium, and none were found. In other words, physical contact between the two strains was needed for wild-type cells to form. It looked as though some kind of gene transfer had taken place, and genetic recombinants were indeed produced. Discovery of the fertility factor (F) In 1953, William Hayes determined that genetic transfer occurred in one direction in the above types of crosses. Therefore, the transfer of genetic material in E. coli is not reciprocal. One cell acts as donor, and the other cell acts as the recipient. This kind of unidirectional transfer of genes was originally compared to a sexual difference, with the donor being termed “male” and the recipient “female.” However, this type of gene transfer is not true sexual reproduction. In bacterial gene transfer, one organism receives genetic information from a donor; the recipient is changed by that information. In sexual reproduction, two organisms donate equally (or nearly so) to the formation of a new organism, but only in exceptional cases is either of the donors changed. MESSAGE The transfer of genetic material in E. coli is not reciprocal. One cell acts as the donor, and the other cell acts as the recipient. Loss and regain of ability to transfer. By accident, Hayes discovered a variant of his original donor strain that would not produce recombinants on crossing with the recipient strain. Apparently, the donor-type strains had lost the ability to transfer genetic material and had changed into recipient-type strains. In his analysis of this “sterile” donor variant, Hayes realized that the fertility (ability to donate) of E. coli could be lost and regained rather easily. Hayes suggested that donor ability is itself a hereditary state imposed by a fertility factor (F). Strains that carry F can donate, and are designated F + . Strains that lack F cannot donate and are recipients. These strains are designated F − . Transfer of F during conjugation Recombinant genotypes for marker genes are relatively rare in bacterial crosses, Hayes noted, but the F factor apparently was transmitted effectively during Figure 7-4 Bacteria can transfer plasmids physical contact, or conjugation. A kind of “infectious transfer” of the F factor seemed to be taking place. We now know much more about the process of conjugation and about F, which is an example of a plasmid that can replicate in the cytoplasm independently of the host chromosome. Figures 7-4 and 7-5 show (circles of how bacteria can transfer plasmids such as F. The F plasmid directs the synthesis DNA), through of pili, projections that initiate contact with a recipient (Figure 7-4 ) and draw it (more...) closer, allowing the F DNA to pass through a pore into the recipient cell. One strand of the double-stranded F DNA is transferred and then DNA replication restores the complementary strand in both the donor and the recipient. This replication results in a copy of F remaining in the donor and another appearing in Figure 7-5 the recipient, as shown in Figure 7-5 . (a) During conjugation, the pilus pulls two bacteria (more...) Hfr strains An important breakthrough came when Luca Cavalli-Sforza discovered a derivative of an F + strain. On crossing with F − strains this new strain produced 1000 times as many recombinants for genetic markers as did a normal F + strain. Cavalli-Sforza designated this derivative an Hfr strain to indicate a high frequency of recombination. In Hfr × F − crosses, virtually none of the F − parents were converted into F + or into Hfr. This result is in contrast with F + × F − crosses, where infectious transfer of F results in a large proportion of the F − parents being converted into F + . Figure 7-6 portrays this concept. It became apparent that an Hfr strain results from the integration of the F factor into the chromosome, as Figure 7-6 pictured in Figure 7-6a . The transfer of E. coli chromosomal markers Now, during conjugation between an Hfr cell and a F − cell a part of the mediated before the entire chromosome is transferred. The chromosomal fragment can then (more...) recombine with the recipient chromosome. Clearly, the low level of chromosomal chromosome is transferred with F. Random breakage interrupts the transfer marker transfer observed by Lederberg and Tatum (see Figure 7-2 ) in an F + × F − cross can be explained by the presence of rare Hfr cells in the population. When these cells are isolated and purified, as first done by Cavalli, they now transfer chromosomal markers at a high frequency, because every cell is an Hfr. Determining linkage from interrupted-mating experiments The exact nature of Hfr strains became clearer in 1957, when Elie Wollman and François Jacob investigated the pattern of transmission of Hfr genes to F − cells during a cross. They crossed Hfr str s a+ b + c+ d + with F − str r a− b − c− d − . At specific time intervals after mixing, they removed samples. Each sample was put in a kitchen blender for a few seconds to disrupt the mating cell pairs and then was plated onto a medium containing streptomycin to kill the Hfr donor cells. This Figure 7-7 Interruptedmating conjugation experiments procedure is called interrupted mating. The str r cells then were tested for the presence of marker alleles from the donor. Those str r cells bearing donor marker alleles must have taken part in conjugation; such cells are called exconjugants. Figure 7-7a shows a plot of the results; azi r, ton r, lac + , and gal + correspond to with E. the a+ , b + , c+ , and d + mentioned in our generalized description of the (more...) experiment. Figure 7-7b portrays the transfer of markers. The most striking thing about these results is that each donor allele first appeared in the F − recipients at a specific time after mating began. Furthermore, the donor alleles appeared in a specific sequence. Finally, the maximal yield of cells containing a specific donor allele was smaller for the donor markers that entered later. Putting all these observations together, Wollman and Jacob concluded that gene transfer occurs from a fixed point on the donor chromosome, termed the origin (O), and continues in a linear fashion. MESSAGE The Hfr chromosome, originally circular, unwinds and is transferred to the F − cell in a linear fashion. The unwinding and transfer begin from a specific point at one end of the integrated F, called the origin or O. The farther a gene is from O, the later it is transferred to the F − ; the transfer process most likely will stop before the farthermost genes are transferred. Wollman and Jacob realized that it would be easy to construct linkage maps from the interrupted-mating results, using as a measure of “distance” the times at Figure 7-8 which the donor alleles first appear after mating. The units of distance in this Chromosome case are minutes. Thus, if b + begins to enter the F − cell 10 minutes after a+ map of Figure begins to enter, then a+ and b + are 10 units apart (Figure 7-8 ). Like the maps 7-7. A linkage map can be based on crossover frequencies, these linkage maps are purely genetic constructions; at the time, they had no known physical basis. (more...) Chromosome circularity and integration of F When Wollman and Jacob allowed Hfr × F − crosses to continue for as long as 2 hours before blending, they found that a few of the exconjugants were converted into Hfr. In other words, an important part of F (the terminal part now known to confer “maleness,” or donor ability), was eventually being transmitted but at a very low efficiency, and it apparently was transmitted as the last element of the linear chromosome. We now have the following map, in which the arrow indicates the process of transfer, beginning with O: However, when several different Hfr linkage maps were derived by interrupted-mating and time-of-entry studies using different, separately derived Hfr strains, the maps differed from strain to strain. At first glance, there seems to be a random reshuffling of genes. However, a pattern does exist; the genes are not thrown together at random in each strain. For example, note that in every case the his gene has gal on one side and gly on the other. Similar statements can be made about each gene, except when it appears at one end or the other of the linkage map. The order in which the genes are transferred is not constant. In two Hfr strains, for example, the his gene is transferred before the gly gene (his is closer to O), but, in three strains, the gly gene is transferred before the his gene. How can we account for these unusual results? Allan Campbell proposed a startling hypothesis: suppose that, in an F + male, F is a small cytoplasmic element (and therefore easily transferred to an F − cell on conjugation). If the Figure 7-9 Circularity of the E. coli chromosome. (a) Through (more...) chromosome of the F + male were a ring, any of the linear Hfr chromosomes could be generated simply by inserting F into the ring at the appropriate place and in the appropriate orientation (Figure 7-9 ). Several conclusions—later confirmed—follow from this hypothesis. 1. The orientation in which F is inserted would determine the polarity of the Hfr chromosome, as indicated in Figure 7-9a . 2. At one end of the integrated F factor would be the origin, where transfer of the Hfr chromosome begins; the terminus at the other end of F would not be transferred unless all the chromosome had been transferred. Because the chromosome often breaks before all of it is transferred and because the F terminus is what confers maleness, then only a small fraction of the recipient cells would be converted into male cells. How, then, might F integration be explained? Wollman and Jacob suggested that some kind of crossover event between F and the F + chromosome might generate the Hfr chromosome. Campbell then came Figure 7-10 Insertion of the F factor into the up with a brilliant extension of that idea. He proposed that, if F, like the chromosome, were circular, then a crossover between the two rings would produce a single larger ring with F inserted (Figure 7-10 ). E. coli chromosome Now suppose that F consists of three different regions, as shown in (more...) Figure 7-10 . If the bacterial chromosome had several homologous regions that could match up with the pairing region of F, then different Hfr chromosomes could be easily generated by crossovers at these different sites. Chromosomal and F circularity were wildly implausible concepts initially, inferred solely from the genetic data; confirmation of their physical reality came only a number of years later. The direct-crossover model of integration also was subsequently confirmed. The fertility factor thus exists in two states: (1) the plasmid state, as a free cytoplasmic element F that is easily transferred to F − recipients, and (2) the integrated state, as a contiguous part of a circular chromosome that is transmitted only very late in conjugation. The word episome (literally, “additional body”) was coined for a genetic particle having such a pair of states. A cell containing F in the first state is called an F + cell, a cell containing F in the second state is an Hfr cell, and a cell lacking F is an F − cell. Today the term plasmid is used to refer to any selfreplicating circular element in the cytoplasm and “episome” is rarely used. R factors A frightening ability of pathogenic bacteria was discovered in Japanese hospitals in the 1950s. Bacterial dysentery is caused by bacteria of the genus Shigella. This bacterium initially proved sensitive to a wide array of antibiotics that were used to control the disease. In the Japanese hospitals, however, Shigella isolated from patients with dysentery proved to be simultaneously resistant to many of these drugs, including penicillin, tetracycline, sulfanilamide, streptomycin, and chloramphenicol. This multiple-drugresistance phenotype was inherited as a single genetic package, and it could be transmitted in an infectious manner—not only to other sensitive Shigella strains, but also to other related species of bacteria. This talent is an extraordinarily useful one for the pathogenic bacterium, and its implications for medical science were terrifying. From the point of view of the geneticist, however, the situation is very interesting. The vector carrying these resistances from one cell to another proved to be a selfreplicating element similar to the F factor. These R factors (for “resistance”) are transferred rapidly on cell conjugation, much like the F particle in E. coli. In fact, these R factors proved to be just the first of many similar F-like factors to be discovered. These elements, which exist in the plasmid state in the cytoplasm, have been found to carry many different kinds of genes in bacteria. Table 7-2 shows some of the characteristics that can be borne by plasmids. Table 7-2 Genetic Determinants Borne by Plasmids Mechanics of transfer Does an Hfr cell die after donating its chromosome to an F − cell? The answer is no (unless the culture is treated with streptomycin). The Hfr chromosome replicates while it is transferring a single strand to the F − cell; this replication ensures a complete chromosome for the donor cell after mating. The transferred strand is replicated in the recipient cell, and donor genes may become incorporated in the recipient’s chromosome through crossovers, creating a recombinant cell. Otherwise, transferred fragments of DNA in the recipient are lost in the course of cell division. We assume that the F − chromosome is also circular, because the recipient F − cell, if it receives the F factor from an F + cell, is readily converted into an F + cell from which an Hfr cell can be derived. The picture emerges of a circular Hfr chromosome unwinding a copy of itself, which is then transferred in a linear fashion into the F − cell. How is the transfer achieved? Electron microscope studies show that Hfr and F + cells have fibrous structures, F pili, protruding from their cell walls, as shown in Figure 7-4 . The F pili facilitate cell-to-cell contact, during which DNA is transferred through pores in the F − . E. coli conjugation cycle We can now summarize the various aspects of the conjugation cycle in E. coli (Figure 7-11 ). We shall review the conjugation cycle in regard to the differences between F − , F + , and Hfr cells, because these differences epitomize the cycle. Figure 7-11 Summary of the various events that take place in the (more...) F − strains do not contain the F factor and cannot transfer DNA by conjugation. They are, however, recipients of DNA transferred from F + or Hfr cells by conjugation. F + cells contain the F factor in the cytoplasm and can therefore transfer F in a highly efficient manner to F − cells during conjugation. Hfr cells have F integrated into the bacterial chromosome, not in the cytoplasm. Chromosomal markers are transferred in a strain of F + cells because, in any population of F + cells, a small fraction of cells (about 1 in 1000) have been converted into Hfr cells by the integration of F into the bacterial chromosome. Because conjugation experiments are usually carried out by mixing from 107 to 10 8 cells consisting of prospective donors and recipients, the population will contain various different Hfr cells derived from independent integrations of F into the chromosome at various different sites. Therefore, when chromosomal markers are transferred by different cells in the population, transfer will start at different points on the chromosome. This results in an approximately equal transfer of markers all around the chromosome, although at a low frequency. This type of F + -mediated transfer is what Lederberg and Tatum observed when they discovered gene transfer in bacteria. Each of the Hfr cells in an F + population with an integrated F factor can be the source of a new Hfr strain if it is isolated and used to start a clone. Hfr strains are derived from a clone of Hfr cells in which a specific integration of F into the bacterial chromosome has taken place. Therefore, all the cells in any given Hfr strain have F integrated into the chromosome at exactly the same point. Hfr populations transfer chromosomal markers to F − cells at a high frequency compared with F + populations, because only a fraction of cells in an F + population have F integrated into the chromosome. Further, in any given Hfr strain, the markers are transferred from a fixed point in a specific order. This also contrasts with F + populations, where the Hfr cells transfer chromosomal markers in no particular fixed order, given that the F factor integrates into the chromosome at different points in different F + cells. In an Hfr × F − cross, the F − is not converted into Hfr or into F + , except in very rare cases, because the Hfr chromosome nearly always breaks before the F terminus is transferred to the F− cell. Recombination between marker genes after transfer Thus far, we have studied only the process of the transfer of genetic information between individuals in a cross. This transfer is inferred from the existence of recombinants produced from the cross. However, before a stable recombinant can be produced, the transferred genes must be integrated or incorporated into the recipient’s genome by an exchange mechanism. We now consider some of the special properties of this exchange event. Genetic exchange in prokaryotes does not take place between two whole genomes (as it does in eukaryotes); rather, it takes place between one complete genome, derived from F − , called the endogenote, and an incomplete one, derived from the donor, called the exogenote. What we have in fact is a partial diploid, or merozygote. Bacterial genetics is merozygous genetics. Figure 7-12a is a diagram Figure 7-12 Crossover between exogenote and endogenote in a merozygote (more...) of a merozygote. A single crossover would not be very useful in generating viable recombinants, because the ring is broken to produce a strange, partly diploid linear chromosome (Figure 7-12b). To keep the ring intact, there must be an even number of crossovers (Figure 7-12c ). The fragment produced in such a crossover is only a partial genome, which is generally lost in subsequent cell growth. Hence, both reciprocal products of recombination do not survive—only one does. A further unique property of bacterial exchange, then, is that we must forget about reciprocal exchange products in most cases. MESSAGE In the genetics of bacteria, we generally are concerned with double crossovers and we do not expect reciprocal recombinants. Gradient of transfer Only partial diploids exist in the merozygote. Some genes don’t even get into the act. To better understand this fact, let us look again at the consequences of gene transfer. Usually, only a fragment of the donor chromosome appears in the recipient, owing to spontaneous breakage of the mating pairs; so the entire chromosome is rarely transferred. The spontaneous breakage can occur at any time after transfer begins, which creates a natural gradient of transfer and makes it less and less likely that a recipient cell will receive later and later genetic markers. (“Later” here refers to markers that are increasingly farther from the origin and hence are donated later in the order of markers transferred.) For example, in a cross of Hfr-donating markers in the order met, arg, leu, we would expect a distribution of fragments such as the one represented here: Note that many more fragments contain the met locus than the arg locus and that the leu locus is present on only one fragment. It is easy to see that the closer the marker is to the origin, the greater the chance it will be transferred in conjugation. The concept of the gradient of transfer is the same as the one described earlier for interrupted matings, except that here we are allowing the natural disruption of mating pairs to occur instead of interrupting the pairs mechanically. Determining gene order from gradient of transfer We can use the natural gradient of transfer to establish the order of genetic markers, provided we select for an early marker that enters before the markers that we are ordering. Let’s see how this works. Suppose that we use an Hfr strain that donates markers in the order met, arg, aro, his. In a cross of an Hfr that is met + arg + aro + his + str s with an F − that is met − arg − aro − his − str r, recombinants are selected that can grow on a minimal medium without methionine but with arginine, aromatic amino acids, and histidine and in the presence of streptomycin. Here we are selecting for recombinants in the F − strain that are met + in a cross in which the met locus is transferred as the earliest marker. We can then score for inheritance of the other markers present in the Hfr by testing on supplemental minimal medium lacking, in turn, one of the required nutrients. A typical result would be: Note how the frequency of inheritance corresponds to the order of transfer. This correspondence is due to the fact that the frequency of inheritance is indicative of the frequency of transfer. For this method to work, it is crucial that it be applied only to genetic markers that enter after the selected marker—in this case, after met. Higher-resolution mapping by recombinant frequency in bacterial crosses Although interrupted-mating experiments and the natural gradient of transfer can give us a rough set of gene locations over the entire map, other methods are needed to obtain a higher resolution between marker loci that are close together. Here we consider one approach to the problem: using the frequency of recombinants to measure linkage. Previous attempts to measure linkage in conjugational crosses were hindered by the failure to understand that only fragments of the chromosome are transferred and that the gradient of transfer produces a bias toward the inheritance of early markers. To measure linkage and to attach any meaning to a calculated map distance, it is necessary to produce a situation in which every marker has an equal chance at being transferred so that the recombinant frequencies are dependent only on the distance between the relevant genes. Suppose that we consider three markers: met, arg, and leu. If the order is met, arg, leu and if met is transferred first and leu last, then we want to set up the situation diagrammed here to calculate map distances separating these markers: Here, we have to arrange to select the last marker to enter, which in this case is leu. Why? Because, if we select for the last marker, then we know that every cell that received fragments containing the last marker also received the earlier markers—namely, arg and met—on the same fragments. We can then proceed to calculate map distance in the classic manner. Rather than using map units, we simply refer to the percentage of crossovers in the respective interval on the map. In practice, this is done by calculating, among the total recovered recombinants, the percentage of recombinants produced by crossovers between two markers. Let’s look at an example. Sample cross In the cross of the Hfr strain just described (met + arg + leu + str s) with an F − that is met − arg − leu − str r, we would select leu + recombinants and then examine them for the arg and met markers. In this case, the arg and met Figure 7-13 Incorporation of a late marker into the F − (more...) markers are called the unselected markers. Figure 7-13 depicts the types of crossover events expected. Note how two crossover events are required to incorporate part of the incoming fragment into the F − chromosome. One crossover must be on each side of the selected (leu) marker. Thus, in Figure 7-13 , one crossover must be on the left side of the leu marker and the second must be on the right side. Suppose that the map distance between each marker is 5 percent recombination. In 5 percent of the total leu + recombinants, the second crossover occurs between leu and arg (Figure 7-13a ); in another 5 percent of the cases, the second crossover occurs between leu and met (Figure 7-13b). We would then expect 90 percent of the selected leu + recombinants to be arg + met + , because the second crossover occurs outside the leu–arg–met interval (Figure 7-13c ) in 90 percent of the cases. We would also expect 5 percent of the leu + recombinants to be arg − met − , resulting from a crossover between leu and arg, and 5 percent of the leu + recombinants to be arg + met − , resulting from a crossover between arg and met. In reality, then, we are simply determining the percentage of the time that the second crossover occurs in each of the three possible intervals. In a cross such as the one just described, one class of potential recombinants requires an additional two crossover events (Figure 7-13d). In this case, the leu + arg − met + recombinants would require four crossovers instead of two. These recombinants are rarely recovered, because their frequency is sharply reduced compared with the other classes of recombinants. Infectious marker-gene transfer by episomes Edward Adelberg’s work led to the discovery of gene transfer at high frequency by episomes. When he began his recombination experiments in 1959, the particular Hfr strain that he used kept producing F + cells, so the recombination frequencies were not very large. Adelberg called this particular fertility factor F′ to distinguish it from the normal F, for the following reasons: 1. The F′-bearing F + strain reverted back to an Hfr strain much more frequently than do typical F + strains. 2. F′ always integrated at the same place to give back the original Hfr chromosome. (Remember that randomly selected Hfr derivatives from F + males have origins at many different positions.) How could these properties of F′ be explained? The answer came from the recovery of an F′ from an Hfr strain in which the lac + locus was near the end of the Hfr chromosome (it was transferred very late). Using this Hfr lac + strain, François Jacob and Adelberg found an F + derivative that transferred lac + to F − lac − recipients at a very high frequency. Furthermore, the recipients that behaved like F + lac + occasionally produced F − lac − daughter cells, at a frequency of 1 × 10 −3. Thus, the genotype of these recipients appeared to be F lac + /lac − . Now we have the clue: F′ is a cytoplasmic element that carries a part of the bacterial chromosome. In fact, it is nothing more than F with a piece of the host chromosome incorporated. Its origin and reintegration can be visualized as shown in Figure 7-14 . This F′ is known as F′ lac, because the piece of host chromosome that it picked up has the lac gene on it. F′ factors have been found carrying many different chromosomal genes and have been named accordingly. For Figure 7-14 example, F′ factors carrying gal or trp are called F′ gal and F′ trp, respectively. Origin and reintegration of the F′ factor, (more...) Because F lac + /lac − cells are Lac+ in phenotype, we know that lac + is dominant over lac − . As we shall see in Chapter 11, the dominant– recessive relation between alleles can be a very useful bit of information in interpreting gene function. Partial diploidy for specific segments of the genome can be made with an array of F′ derivatives from Hfr strains. The F′ cells can be selected by looking for the infectious transfer of normally late genes in a specific Hfr strain. Some F′ strains can carry very large parts (up to one-quarter) of the bacterial chromosome; if appropriate markers are used, the merozygotes generated can be used for recombination studies. MESSAGE During conjugation between an Hfr donor and an F − recipient, the genes of the donor are transmitted linearly to the F − cell, through the bacterial chromosome, with the inserted fertility factor transferring last. In the course of conjugation between an F + donor carrying an F′ plasmid and an F − recipient, a specific part of the donor genome may be transmitted infectiously to the F − cell, through the plasmid. The transmitted part was originally adjacent to the F locus in an Hfr strain from which the F + was derived. Copyright © 2000, W. H. Freeman and Company Bookshelf ǀ NCBI ǀ NLM ǀ NIH Help ǀ Contact Help Desk ǀ Copyright and Disclaimer Sample cross Infectious marker-gene transfer by episomes