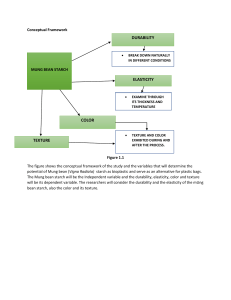
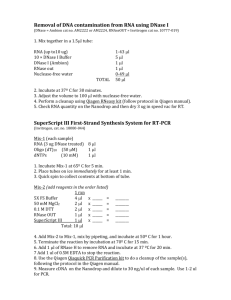
Supplementary Information
advertisement

Supplementary Material for Chemical Communications This journal is © The Royal Society of Chemistry 2003 Supplementary Information A/A 0 at 260 nm 1.3 1.2 1.1 1 0 10 20 30 40 Time / min 50 60 Fig. S1 A time course of absorbance increase of the ligated nucleo-nanocages 1-3, due to the enzymatic action of DNase I. [nucleobase] = 30 µM, enzyme, 30 unit in 330 µL of 20 mM Tris-HCl buffer (pH 7.5, containing 0.5 M NaCl, 1 mM CaCl2, and 1 mM MgCl2) at 37 °C. Absorption changes were monitored at 260 nm. As shown in Figure S1, the ligated nucleo-nanocages are readily digested with DNase I under the standard condition. Molecular weight of DNase I is 29 kDa, which is comparable to these of exonuclease III (31 kDa) and mung bean nuclease (39 kDa). Therefore, it is reasonable to assume that the observed resistance towards exonuclease III and mung bean nuclease are ascribed to the absence of end structures, but not to the steric hindrance.