emi1136AppendixS1+and+TableS1
advertisement

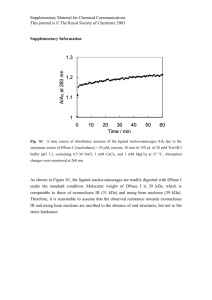
1 Supplementary material 2 3 Appendix S1. 4 Total RNA isolation 5 Total RNA (totRNA) was isolated from cultures of B. cepacia strain 2a and its mutants 6 grown in MSM supplemented with 2 mM succinate and 2.26 mM 2,4-D using a silica- 7 magnetite magnetisable solid-phase support, produced at the University of Kent. 8 9 Frozen cell pellets were thawed on ice, resuspended in 100µl TE buffer containing 10 lysozyme (400µg/ml) and incubated for 5 minutes at 20°C. Then, 200µl of lysis buffer (50 11 mM Tris-HCl pH8; 25 mM EDTA pH8; 4 M guanidine thiocyanate; 0.01% Triton-X-100; 1% 12 β-mercaptoethanol) was added and the mixtures vortexed for 5 seconds. Subsequently, 13 300 µl of isopropanol was added and the resultant volume transferred to a sterile 1.5 ml 14 Eppendorf tube, containing 2 mg of silica-magnetite. The mixture was incubated with 15 gentle agitation for 5 minutes at 20°C, the silica-magnetite immobilised using a magnetic 16 stand (Promega Ltd.) and the supernatant removed and discarded. The silica-magnetite 17 was then washed with 200 μl of cold 80% (v/v) aqueous ethanol, immobilised 18 ‘magnetically’ the supernatant removed and the silica-magnetite left to air dry for 5 19 minutes. RNA was then eluted from the support by addition of 70 μl of nuclease free water 20 followed by incubation with gentle agitation for 1 minute at room temperature. The support 21 was then immobilised and the RNA-containing supernatant transferred to a sterile 1.5 ml 22 Eppendorf tube and the elution process repeated and the eluates combined. 1 µl of the 23 product was removed for quantification and purity measurement (OD260/280). RNasin® 24 (Promega Ltd.) was added to the remaining sample to a final concentration of 1U/μl and 5 25 μl of the solution mixed with 2 μl of 10X RNA loading buffer (Sigma Ltd) and loaded onto a 26 2% (w/v) agarose gel (Hi-PureTM low EEO agarose) for physical quality assurance. 27 Undegraded totRNA was confirmed by the state of intactness of the 23 and 16S rRNA 28 bands. 29 Prior to quantitative reverse transcription real time-PCR, totRNA samples were treated 30 with DNase I to remove any contamining DNA in the following manner: 2 μg of RNasin ® 31 containing totRNA was placed in a sterile 0.5 ml Eppendorf tube containing 1μl of 10X 32 DNase I reaction buffer (200 mM Tris-HCl pH 8.4, 20mM MgCl2, 500 mM KCl), 1 U DNase 33 I (Promega Ltd.) and the volume made up to 10 μl with nuclease free water. The reaction 34 mixture was incubated for 30 minutes at 37°C and stopped by addition of 1 μl of RQ1 35 DNase stop solution (20 mM EGTA pH 8) and heating to 65°C for 10 minutes. 36 Subsequently, 2 μl of DNase I treated total RNA was mixed with 2 μl of RNA loading buffer 37 (50% (v/v) glycerol; 1 mM EDTA pH 8; 0.5% bromophenol blue; 1 μg/ml ethidium bromide) 38 and loaded onto a 2 % (w/v) agarose gel to check for contaminating DNA. Successful 39 removal of DNA was also confirmed by PCR amplification of DNase I-treated totRNA. The 40 remaining sample was stored at -196°C until required. 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 Table 2. Primers used to construct open- and closed-channel B. cepacia strain 2a mutants 57 Mutant name 58 (genotype) Sequences (5’ – 3’) Product size (bp) 59 60 Open-channel Fw- CGGAATTCCGTTCGGCTTACTGCGGCAAAG 651 EcoRI 61 (Glu162Ala) Rw- CGGATCCCGTCGCGACGAACTTGAGTAAG BamHI 62 Mut- TCGATTGGGCGCGCTGGCCCTTCAGTGC GAG 63 64 Closed-channel Fw- CGGAATTCCGTTCGGCTTACTGCGGCAAAG 651 EcoRI 65 (Glu162Asp) Rw- CGGATCCCGTCGCGACGAACTTGAGTAAG BamHI 66 Mut- CGATTGGGCGCGGATGGCCCTTCAGTGC GAG 67 68 Mutations were generated by amplification of pRP15c with primers designed using 69 PrimerX (available at http://bioinformatics.org/primerx/) according to Ke and Madison 70 (1997) and were modified so as to possess EcoRI and BamHI restriction sites (underlined) 71 allowing products to be used in subsequent cloning. The nucleotide changes to introduce 72 alanine (Ala) and aspartate (Asp) in place of a glutamate (Glu) are underlined in the 73 mutagenic primers (Mut). GAG, GCT and GAT represented the trinucleotides encoding 74 Glu, Ala and Asp, respectively. 75