Sequencing Of The Entire NOD2/CARD15 Gene: Analysis
advertisement

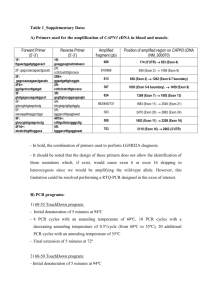
Supplemental table 1: Primers used in the sequencing of the NOD2/CARD15 gene Exon Annealing Amplicon size temperature(C) (base pairs) 1. TTGTGCCAGAATTGCTTG AAGGGTAGAATAAGCTCTGGG 60 503 2. TCTGAGGCTAGAACCATGG TGAGGACAAATCAGTCTTGG 57 622 3. GACCCTTTATTCTGGATGGAAG CGGTACAGATAATGAGAGTTTGG 60 486 4(1) TGCTCTCCTATCCCTTCAG TCAGAGAAGCCCTTGAGG 57 784 4(2) GAAGTACATCCGCACCGAG GAAGGCTGCTGTGATCTG 58 690 4(3) CCAGGCAACTCACCAATG AAGGGAAGGGATCTGGG 60 667 5,6 CACTTCAGGGATGAATGAAAG GCATTAGAGAACCCCTGC 58 589 7. GTCTTCAATGCTTTCTTCCTG TCTTGTCAAATGGACTCCAG 57 834 8. AAGTCTGTAATGTAAAGCCAC CCCAGCTCCTCCCTCTTC 60 281 9. GAGCACCGCAATCAATTAG CACTCAATCATCCACCTTTG 60 407 10. TTCTTTATCCATGAGTTTGGG CTTTATTGGTTACCTTCACTTC 56 401 11. GAAGAGAGACGGTTACATTTCAC CATTCTTCAACCACATCCC 60 522 12(1) TAAAAACAGCCCTGACTTCC AATTGTCTTGGGGAACAAAC 60 883 12(2) ATTCAGAATATTAGTGACCTCAGC ATGTTGGTCAGGTTGGTC 58 866 1 Supplemental table 2: SNPs identified in sequencing of the NOD2/CARD15 gene SNP SNP description Patient rs number frequency Exon number / base change 1 upstream 1 rs5743264 Exon 1 promoter T/C 2 5 UTR(-59) 16 rs5743266 Exon 1 G/A 3 intronic 3 rs2076753 Exon 2 promoter G/T* 4 Ser178Ser 13 rs2067085 Exon 2 C/G 5 Ala211Ala 1 rs5743269 Exon 3 C/T 6 Pro268Ser (SNP 5) 14 rs2066842 Exon 4(1) C/T 7 Arg459Arg (SNP 13 rs2066843 Exon 4(2) C/T 12 rs1861759 Exon 4(2) T/G 1 ** Exon 4(2) C/T rs2066844 Exon 4 (3)C/T ** Exon 4 end + 10 bases 6) 8 Arg587Arg (SNP 7) 9 10 Arg702Trp (SNP 8) 5 11 2 A/C 12 Met863Val 2 ** Exon 6 A/G 13 Gly908Arg 2 rs2066845 Exon 8 G/C 4 rs5743291 Exon 9 G/A 10 ** Exon 10 (post exonic (SNP12) 14 Val955Ile 15 +1) T/A 16 Leu1007fsincC 1 rs2066847 Exon 11 c+/- 2 (SNP13) 17 3 UTR 12 rs3135499 Exon 12(1) A/C 18 3 UTR 12 rs3135500 Exon 12(2) G/A Legend: * This is described in Lesage35 (and here) as an Exon 2 promoter mutation, numbered from the ATG in the 2nd exon that was thought to be the start of the NOD2/CARD15. It is actually in intron one/two. ** No rs number listed in Ensembl DNA was extracted from whole blood using the salting out technique.48 Direct sequencing was performed on the 7900 HT sequence detection system (Applied Biosystems, Foster City, Ca, USA) by the Technical Services section of the MRC Human Genetics Unit, Edinburgh. 24 Caucasian patients (<16 years at diagnosis) with CD were studied with the number of patients variants were found in listed in the table. DNA sequence was analysed using Sequencher v4.5 (Gene Codes Corporation, Ann Arbor, MI, USA). 3 Supplemental table 3: Demographics of patients involved in the study Cohort 1 Cohort 2 175/145 137/206 11.08 28.25 IQR (8.55-12.92) IQR(21.00-42.00) Current Smoker 7/315 (2.2%) 94/333 (28.2%) Family history of 107/315 (34.0%) 66/343 (19.2%) 313 (97.8%) 329/331 (99.4%) Sex (M/F) Median Age at diagnosis(y) IBD Caucasian (%) Legend: Cohort 1 320 IBD patients <16 years at diagnosis; Cohort 2 343 CD patients from adult cohort. Patients were recruited to the study from the three tertiary paediatric gastroenterology clinics in Scotland and the Western General Hospital, Edinburgh. A diagnosis of IBD was based on standard criteria.49 Patient data collection and disease phenotyping was performed as previously described.19;20;22 Informed written consent was obtained from parents and patients prior to participation in the study. Ethical approval was obtained from the local research ethics committees in all the participating hospitals. 4 Supplemental table 4: Case-control analysis in the different disease groups Disease 863M/V p value cf. 955V/I variant p value cf. group carriage HC carriage HC HC 1/267 (0.37%) 41/253 (16.2%) CD children 2/207 (0.97%) 0.58 29/202 (14.4%) 0.29 CD adults 5/313 (1.6%) 0.22 36/305 (11.8%) 0.13 Both CD 7/520(1.35%) 0.27 65/507 (12.8%) 0.20 UC 0/80 - 12/78 (15.8%) 0.86 IC 0/30 - 12/29 (41.4%) 0.001 Legend: HC=healthy controls; CD=Crohn’s disease; UC=ulcerative colitis; IC=indeterminate colitis. The numbers of carriers of the variant form of the SNP analysed is compared to the number of patients successfully genotyped e.g. there were 273 healthy controls of which 267 were successfully genotyped for the 863M/V SNP. TaqMan (Applied Biosystems, Foster City, CA, USA) was used to genotype 955V/I (rs5743291) and 863M/V. Minitab version 13 (Release 13.20, Minitab Inc., State College, PA, USA) statistical package was used to analyse genotype associations using chi squared or Fishers exact test where appropriate. A total of 253 healthy populations controls (101 healthy controls and 152 blood donors) were used for casecontrol analysis. 5