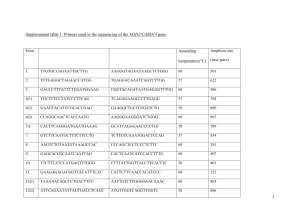
Table 2: Primers used for the amplification of CAPN3 cDNA in blood
advertisement

Table 1_Supplementary Data: A) Primers used for the amplification of CAPN3 cDNA in blood and muscle. Forward Primer (5’-3’) Reverse Primer (5’-3’) 1F: ttgaactggatgtggacact 1R: gcaggcagtcatctataacc 2R: ccttctcatctttgtccaca 2R6+: ggaatgattgtccggtc 2R: ccttctcatctttgtccaca 3R: gcgttgtacaggaagaagtc 4R: tctcgtagctgttgatggtg 5R: tgggacctttgaggtaaat 4R16+: cttttgcttatcagggcttg 5R: tgggacctttgaggtaaat 2F: gagccaacagaactgacatc 2F: gagccaacagaactgacatc 2F6+: ggctgctccattgatgat 3F gtatgagacaagaatggcct 4F: gtaccgtctgaagctcctg 5F: aacaagattaaggcctggc 4F: gtaccgtctgaagctcctg 4F16+: atcatcttcgtttcggaca Amplified fragment (pb) Position of amplified region on CAPN3 cDNA (NM_000070) 680 174 (5’UTR) 853 (Exon 4) 810/666 650 (Exon 2) 1459 (Exon 9) 613 650 (Exon 2) 1262 (Exon 6-7 boundary) 367 1093 (Exon 5-6 boundary) 1459 (Exon 9) 624 1269 (Exon 7) 1892 (Exon 13) 863/845/731 1683 (Exon 11) 2545 (Exon 21) 393 2470 (Exon 20) 2862 (Exon 24) 508 1683 (Exon 11) 2208 (Exon 16) 753 2110 (Exon 16) 2862 (3’UTR) - In bold, the combination of primers used to perform LGMD2A diagnosis. - It should be noted that the design of these primers does not allow the identification of those mutations which, if exist, would cause exon 6 or exon 16 skipping in heterozygosis since we would be amplifying the wild-type allele. However, this limitation could be resolved performing a RTQ-PCR designed in the exon of interest. B) PCR programs: 1) 60-55 TouchDown program: - Initial denaturation of 5 minutes at 94ºC - 4 PCR cycles with an annealing temperature of 60ºC, 10 PCR cycles with a decreasing annealing temperature of 0.5º/cycle (from 60ºC to 55ºC), 20 additional PCR cycles with an annealing temperature of 55ºC - Final extension of 5 minutes at 72º 2) 60-50 TouchDown program: - Initial denaturation of 5 minutes at 94ºC - 4 PCR cycles with an annealing temperature of 60ºC, 10 PCR cycles with a decreasing annealing temperature of 1º/cycle (from 60ºC to 50ºC), 20 additional PCR cycles with an annealing temperature of 50ºC - Final extension of 5 minutes at 72º 3) Extaq Program: - Initial denaturation of 5 minutes at 94ºC - 30 cycles: 98ºC/10sec; 60ºC/30sec; 72ºC/1min30sec - Final extension of 7 minutes at 72º