Kate Sergeant
advertisement

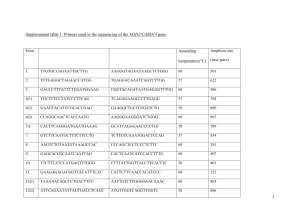
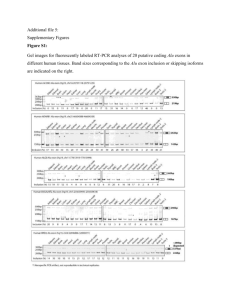
Mutation scanning in Marfan syndrome using High Resolution Melt analysis Kate Sergeant, Northern Genetics Service, Newcastle upon Tyne Marfan syndrome Autosomal dominant, 1 in 5 000 – 1 in 10 000 Connective tissue disorder Affects ocular, skeletal & cardiovascular systems – risk of sudden death FBN1 chr 15, 65 exons 350 kDa extracellular matrix protein Fibrillin FBN1 mutations Over 600 reported mutations (UMD-FBN1) Most mutations are unique Most pathogenic mutations are missense affecting cysteine residues Mutation analysis of FBN1 exons detects ~80% Identifying a mutation gives a definitive diagnosis – cardiological screening to those at risk Aims Set up an assay for mutation scanning in FBN1 Using the LightScanner High Resolution Melt (HRM) analysis system for heteroduplex detection Validate this method using positive controls Test Marfan syndrome patients for FBN1 mutations LightScanner HRM system + variant Temperature D Fluorescence Fluorescence HRM analysis variant Temperature FBN1 assay design Validation with positive controls Exon Nucleotide change Exon Nucleotide change 2 c.247+1G>A 33 c.4139G>A 3 c.306T>C 34 c.4270C>G 5 c.443-35A>G 35 c.4408T>C 6 c.718C>T 37 c.4588C>T 7 c.772C>T 39 c.4942+3_4942+9del7 9 c.1122delT 43 c.5297-2A>G 14 c.1793insTT 45 c.5671+28dupT 15 c.1875T>C 46 c.5672-63G>T 16 c.2023_2026delTTTG 47 c.5816G>A 21 c.2559C>A 53 c.6594C>T 22 c.2684_2689del6 54 c.6617-21A>T 28 c.3511T>C 55 c.6817A>G 29 c.3609_3610ins13 56 c.6888G>A 31 c.3963A>G 57 c.7204+63C>A 32 c.4038C>G 63 c.7852G>A Results Exon 2 Exon 29 c.247+1G>A het c.3609_3610ins13 het Exon 43 Exon 57_2 c.7204+63C>A het c.5297-2A>G het Results Exon Nucleotide change Exon Nucleotide change 2 c.247+1G>A 33 c.4139G>A 3 c.306T>C 34 c.4270C>G 5 c.443-35A>G 35 c.4408T>C 6 c.718C>T 37 c.4588C>T 7 c.772C>T 39 c.4942+3_4942+9del7 9 c.1122delT 43 c.5297-2A>G 14 c.1793insTT 45 c.5671+28dupT 15 c.1875T>C 46 c.5672-63G>T () 16 c.2023_2026delTTTG 47 c.5816G>A 21 c.2559C>A 53 c.6594C>T 22 c.2684_2689del6 54 c.6617-21A>T 28 c.3511T>C 55 c.6817A>G 29 c.3609_3610ins13 56 c.6888G>A 31 c.3963A>G 57 c.7204+63C>A 32 c.4038C>G 63 c.7852G>A Identified? Identified? () Exons 46 and 54 – false negatives? Exon 46 c.5672-63G>T het wild type c.5672-63G>T het wild type ? Exon 45 – false negative Exon 45 c.5671+28dupT het Exon 45 – larger sample number c.5671+28dupT het False positives 22 false positives were encountered Problem with archived DNA and different extraction methods Reduce this by Standardising extraction methods Dilute DNA samples in a common buffer Double reaction volume Summary of validation 28 positive controls tested 1 “true” false negative 22 false positives Sensitivity ~ 96% Specificity ~ 94% Patient panel 6 patients tested so far Correctly identified 12 SNPs Reduced number of false positives Specificity ~98% Conclusions + Sensitive + Quick + Low cost False positives Different DNA samples Some user variability Suitable scanning technique for a large gene Acknowledgements All in the Newcastle laboratory David Bourn Claire Healey, Val Wilson & Danny Routledge Salisbury laboratory – Catharina Yearwood