BCB544-2007-Lab8_Key..
advertisement

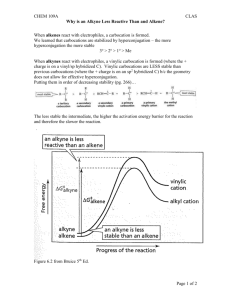
BCB 444/544X Lab 8 Answer Key RNA Secondary Structure Prediction 1. In the color coding scheme, which color means that the base-pair has the highest probability? Which color corresponds to the lowest probability? Red is used for the highest probability and black for the lowest probability. 2. Are there similarities between the structures predicted by mfold and RNAfold? Yes, I thought these structures looked almost the same. The only tricky part was that they were displayed in different orientations by the two servers, which made comparing them a bit more difficult. 3. How does the predicted structures compare to the structure shown above? I saw many differences between the predicted structures and the one shown on the lab. 4. There are 12 questions in the exercise. Submit answers to all 12. 1. List three non-canonical base pairs identified on the web page, besides a G-U wobble base pair. Some choices here are: AC reverse Hoogsteen AC wobble AU reverse Hoogsteen GA imino GG N7-imino Plus lots more 2. What is the computed free energy for this RNA structure? -3.90 kcal/mol 3. Click on the link that says "png" to get a better picture of the structure. How many bases (non-paired) are in the loop in this structure? 4 Note – in both of the structures shown from this server (shown in the answers to #4 and #7 below), there is no small loop and big loop. The “big loop” is not a loop at all, just two free ends that the program unfortunately draws to look a lot like a loop. 4. Save the png file to disk and send a copy to the course instructor. Here is my png file: 5. What is the computed free energy for this RNA structure? -8.10 kcal/mol 6. Click on the link that says "png" to get a better picture of the structure. How many bases (non-paired) are in the loop in this structure? 6 7. Save the png file to disk and send a copy to the course instructor. Here is my png file: 8. Which of these two structures is more likely to exist under physiological circumstances, given no additional constraints? The second structure is more likely because it has a lower energy. 9. How many stems are predicted? 4 stems are predicted. 10. List each of their computed free energy values. Stem Stem Stem Stem 1 = -15.2 kcal/mol 2 = -10.9 kcal/mol 3 = -10.5 kcal/mol 4 = -9.3 kcal/mol 11. Continue to scroll down the page and look at the predicted structure diagram. What is the total free energy of the structure? -28.9 kcal/mol 12. Does this structure that has been predicted from sequences agree well with the known structure of tRNAs? The predicted structure has about the right shape and features of the known structure of tRNAs. 5. Are there any similarities between the HIV and EIAV RRE’s? There is not really any similarity in the predicted structures that I can see, other than possibly both structures have at least one fairly long base-paired region followed by a large bulge. 6. Are there any similarities between the HIV and EIAV RRE structures from mfold? At this point, the structures look more similar than before, but they are still not very close. 7. How does the structure predicted by RNAalifold compare to the mfold structures? At this point, I thought that a portion of the HIV-1 RRE structure from mfold looked fairly similar to a portion of the RNAalifold predicted structure. Specifically, the long hairpin loop with a bulge in the middle of it. However, this is still only very general structural similarity, not obviously similar structures.