pola27182-sup-0001-suppinfo
advertisement

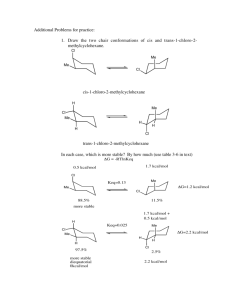
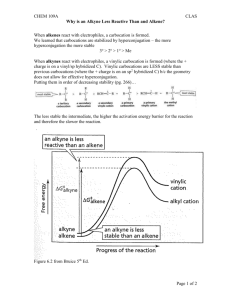
Supporting Information Hyperbranched Aliphatic Polyether Esters by Ring-opening Polymerization of Epoxidized 2-Hydroxyethyl methacrylate Xinhua Huang,1 Min Seon He,1 Jin-Wook Yoo,2 Jin-Seok Choi,3 Il Kim*,1 List of contents 1. Figure S1 Preparation of 2-hydroxyethyl 2-methyloxirane-2-carboxylate (HEMOC). 2. Figure S2 One of the transesterification was occurred between the OH groups and the ester of terminal HEMA units.。 3. Figure S3 Photographic images of the pHEMOCs (H1-H6) changing from sticky liquid to neat solid along with the increase of molecular weight. 4. Figure S4 Variation of radius of gyration according to molecular weight of pHEMOC calculated by molecular dynamics simulation using Forcite module in Accelrys Materials Studio®. 5. Table S1 Collected data from molecular dynamics calculations by using Forcite and Amorphous Cell module in the Materials Studio software (Version 4.3). 6. Figure S5 1H NMR spectra of pHEMOCs: H1–H4 (left) in CDCl3 and H5 and H6 in DMSO-d6. 7. Figure S6 13C, 1H-HSQC spectrum of pHEMOC (H1) in CDCl3 at 25 °C. 8. Figure S7 1H,1H-COSY spectra of pHEMOC (H1) in CDCl3 at 25 °C. 9. Figure S8 1H NMR spetra of pHEMOC (H3) in D2O at different degradation time at 37 ˚C under neutral (pH 7.4) condition. Figure S1 Preparation of 2-hydroxyethyl 2-methyloxirane-2-carboxylate (HEMOC). Figure S2 One of the transesterification was occurred between the OH groups and the ester of terminal HEMA units. Figure S3 Photographic images of the pHEMOCs (H1-H6) changing from sticky liquid to neat solid along with the increase of molecular weight. Figure S4 Variation of radius of gyration according to molecular weight of pHEMOC calculated by molecular dynamics simulation using Forcite module in Accelrys Materials Studio®. Table S1 Collected data from molecular dynamics calculations by using Forcite and Amorphous Cell module in the Materials Studio software (Version 4.3). Property Molecular weight Total enthalpy Total energy Temperature Pressure Volume Density Solubility parameter Bulk modulus (Voigt) Shear modulus (Voigt) Valence energy Bond Angle Torsion Inversion Non-bond energy Hydrogen bond van der Waals Electrostatic Unit g/mol kcal/mol kcal/mol K GPa Å3 g/cm3 (cal/cm3)1/2 GPa GPa kcal/mol kcal/mol kcal/mol kcal/mol kcal/mol kcal/mol kcal/mol kcal/mol kcal/mol Amorphous cell bearing 10 pHEMOC molecules 573 11810.4 4394.6 539.6 8.135 6336.7 1.500 10.31 5.25 2.39 1535.1 495.0 824.1 203.7 12.3 1526.6 -68.7 778.9 816.4 1010 19764.4 7989.2 566.1 7.315 11188.6 1.500 11.94 2.48 2.01 2869.1 901.7 1589.4 357.2 20.8 2709.7 -127.4 1292.5 1544.6 3055 21905.3 19928.8 513.16 0.288 47691.1 1.064 9.63 4.16 2.04 7461.0 2713.7 3832.9 843.5 70.9 6002.1 -145.8 1184.7 4963.3 10364 72687.5 67384.1 510.35 0.221 166783.6 1.032 9.38 3.17 1.39 24933.8 9004.1 12716.3 3007.1 206.3 20814.6 -365.5 3916.6 17263.5 Figure S5 1H NMR spectra of pHEMOCs: H1–H4 (left) in CDCl3 and H5 and H6 in DMSO-d6. Figure S6 13C, 1H-HSQC spectrum of pHEMOC (H1) in CDCl3 at 25 °C. Figure S7 1H,1H-COSY spectra of pHEMOC (H1) in CDCl3 at 25 °C. Figure S8 1H NMR spetra of pHEMOC (H3) in D2O at different degradation time at 37 ˚C under neutral (pH 7.4) condition.