PCR METHODS
advertisement

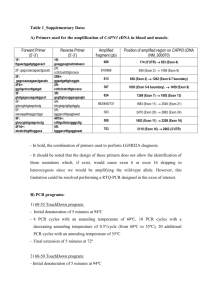
PCR METHODS Ligation-mediated PCR for the analysis of atypical TCRD gene rearrangements Ligation-mediated PCR (LM-PCR) was developed based on the principle of the Universal GenomeWalker Kit (Clontech; Palo Alto, CA). Aliquots of 1 µg high molecular weight DNA were digested with blunt end restriction enzymes (EcoRV, DraI, PvuII and StuI,) and 50 M of an adaptor was ligated to both ends of the restriction fragments. The adaptor was prepared by hybridization of two oligonucleotides: A-sense 5’-GTA ATA CGA CTC ACT ATA GGG CAC GCG TGG TCG ACG GCC CGG GCT GGT and A-reverse 5’-ACC AGC CC-NH2. The amine group blocks the extension of the 3’ end of the reverse oligonucleotide, thereby preventing formation of an AP1 binding site on non-specific adaptor-ligated restriction fragments and adaptor duplexes. The ligation products were subjected to two rounds of PCR with nested adaptor-specific primers AP1 and AP2 and three sets of TCRD specific primers: for the TRDJ1 region: J1(+102) and J1(+62), for the TRDD3 region: D3(+133) and D3(+77) and for the TRDV1 region: V1(-275) and V1(-69). First round PCR of 50 µl total volume was performed with: 1 µl of the ligation products, 10 pmol of the AP1 primer and the J1(+102), D3(+133), or V1(-275) primer, 10 nmol each of dATP, dCTP, dGTP, dTTP (Applied Biosystems, Foster City, CA), 1 µl Advantage Genomic Tth Polymerase Mix, and a Tth PCR buffer (Clontech) including 1.1 mM Mg(OAc) 2 . After the initial denaturation at 94C for 3 min., 7 cycles consisting of 94C for 25 sec. and 72C for 3 min. followed by 32 cycles consisting of 94C for 25 sec. and 67C for 3 min. were performed, with a final extension step at 67C for 7 min. The second round PCR was performed with 1 µl of the first round PCR product, 10 pmol of the AP2 primer and the J1(+62), D3(+77) or V1(-69) primer. Without initial denaturation, 7 cycles consisting of 94C for 25 sec. and 72C for 3 min., followed by 32 cycles consisting of 94C for 25 sec. and 67C for 3 min. were performed and followed by a final extension step at 67C for 7 min. The amplification products were analyzed in a 2% agarose gel. The bands of the leukemic sample that differed in size from those in peripheral blood of healthy individuals, used as a germline control, were excised from the gel and purified using the QIAquick gel extraction kit (Qiagen, Hilden, Germany) and subjected to direct nucleotide sequencing performed by a commercial sequencing facility (AGOWA; Berlin, Germany). RT-PCR and Competitive RT-PCR Total RNA was isolated using RNAeasy columns as described by the manufacturer (Qiagen). A total of 1 µg of RNA was converted to cDNA in a total volume of 20 µl containing RTbuffer (20 mM Tris; 25mM KCL), 1.25 mM MgCl2 (Applied Biosystems; Foster City, Ca, USA), 5 mM DTT (Invitrogen; Groningen, The Netherlands), 5 µM random hexamers (Amersham Pharmacia Biotech, UK), 1 U RNAsin (Promega Corp., Madison, WI, USA), 10 U Superscript RT enzyme (Applied Biosystems) and 1mM dNTPs (Amersham Pharmacia Biotech, UK). RT-PCR of 50 µl total volume was performed with: 1 l cDNA, 10 pmol of each primer, 10 nmol each dATP, dCTP, dGTP, dTTP (Applied Biosystems), 1.5 U AmpliTaq Gold polymerase and a PCR Buffer (Applied Biosystems) including 10 mM TrisHCl, pH 8.3, 50 mM KCl, 2.5 mM MgCl2 and 0.001% (w/v) gelatine. The following primers were used: 5’ primers: BCLf268 specific for the first exon of BCL11B, or BCLf624 specific for the second exon; and 3’ primers: BCLr1029 specifc for the fourth exon of BCL11B, C1(+96) specific for the first exon of the constant region of the TCRD locus and C4(+78) specific for the fourth exon of the TRDC region. After the initial denaturation at 94C for 5 min., 40 cycles consisting of 94C for 1 min., 60C for 1 min. and 72C for 2 min. were performed, followed by the final extension step at 72C for 5 min. Competitive RT-PCR was performed under the same conditions using one 5’ primer: BCLf624 and two 3’ primers: BCLr1029 and C1(+96). As controls brain tissue, 5 T-cell samples obtained from peripheral blood of healthy individuals and the TALL-104 cell line (ATCC) were used. Real-time quantitative PCR Expression levels of wild-type BCL11B and the BCL11B-TCRDfusion mRNA were determined by real-time quantitative PCR using the ABI PRISM 7700 Sequence Detector TaqMan (Applied Biosystems). Two duplex vectors were constructed, containing a fragment of the wild-type BCL11B or BCL11B-TCRD fusion product and, as a reference, a fragment of the 2-microglobuline gene (2MG). The 2MG was cloned first in the T-A acceptor site and subsequently the BCL11B or BCL11B-TCRD were cloned into the EcoRV restriction site of the pCR2.1 TOPO TA Vector (Invitrogen). The vectors were linearized and the number of copies was determined based on the DNA concentration, measured by spectrophotometry and confirmed by a quantitative gel electrophoresis. Standard dilutions of the vector from 107 to 101 copies were prepared. In brief, PCR amplification of a total volume of 50 µl was performed with 2 µl of cDNA, 25 pmol of each primers, 5 pmol of 6FAM-TAMRA probe and the TaqMan PCR Core Reagent Kit (Applied Biosystems). The kit includes 1.5 U AmpliTaq Gold polymerase, 4 mM MgCl2 PCR buffer with 10 nmol of ATP, CTP, GTP and UTP, and 0.5 U AmpErase Uracil N-glycosylase, to prevent a carry-over contamination. The Uracil Nglycosylase was activated by an initial incubation at 50ºC for 2 min., followed by denaturation at 95C for 10 min to activate the AmpliTaq Gold polymerase. Subsequently 45 two-step cycles consisting of 95C for 30 sec and 62C for 1 min. in case of 2MG or 64C for 1 min. in case of both BCL11B and BCL11B-TCRD, were performed. The BCL11B and BCL11B-TCRDamplification products, obtained with the 5’ primer BCLf624 and the 3’ primers: BCLr1029 or C1(+96) respectively, were detected using a probe specific to the second exon BCL11B. The 2MG reference was amplified using the 5’ primer 2MGf41, located in exon 1 and the 3’ primer B2MGb372, located in exon 3, in combination with an exon 2 specific probe.