Supplementary Table S1 (docx 20K)
advertisement
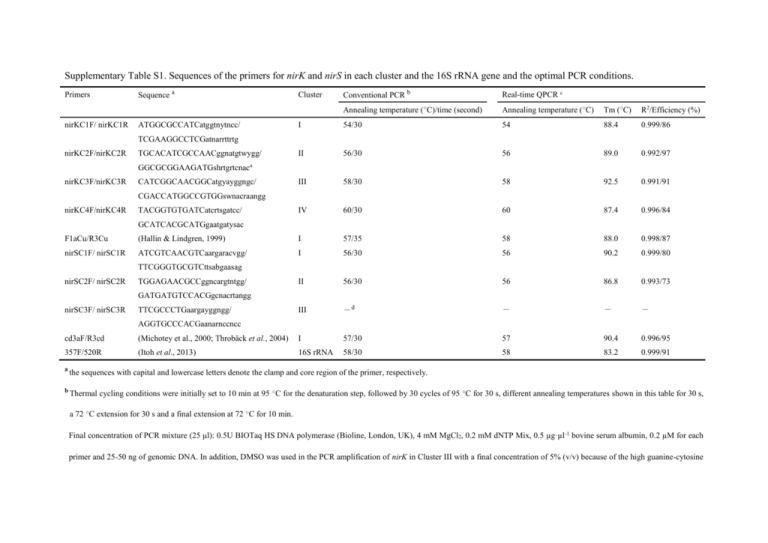
Supplementary Table S1. Sequences of the primers for nirK and nirS in each cluster and the 16S rRNA gene and the optimal PCR conditions. Primers nirKC1F/ nirKC1R Sequence a ATGGCGCCATCatggtnytncc/ Conventional PCR b Real-time QPCR c Annealing temperature (○C)/time (second) Annealing temperature (○C) Tm (○C) R2/Efficiency (%) I 54/30 54 88.4 0.999/86 II 56/30 56 89.0 0.992/97 III 58/30 58 92.5 0.991/91 IV 60/30 60 87.4 0.996/84 Cluster TCGAAGGCCTCGatnarrttrtg nirKC2F/nirKC2R TGCACATCGCCAACggnatgtwygg/ GGCGCGGAAGATGshrtgrtcnaca nirKC3F/nirKC3R CATCGGCAACGGCatgyayggngc/ CGACCATGGCCGTGGswnacraangg nirKC4F/nirKC4R TACGGTGTGATCatcrtsgatcc/ GCATCACGCATGgaatgatysac F1aCu/R3Cu (Hallin & Lindgren, 1999) I 57/35 58 88.0 0.998/87 nirSC1F/ nirSC1R ATCGTCAACGTCaargaracvgg/ I 56/30 56 90.2 0.999/80 II 56/30 56 86.8 0.993/73 III -d - - - TTCGGGTGCGTCttsabgaasag nirSC2F/ nirSC2R TGGAGAACGCCggncargtntgg/ GATGATGTCCACGgcnacrtangg nirSC3F/ nirSC3R TTCGCCCTGaargayggngg/ AGGTGCCCACGaanarnccncc cd3aF/R3cd (Michotey et al., 2000; Throbäck et al., 2004) I 57/30 57 90.4 0.996/95 357F/520R (Itoh et al., 2013) 16S rRNA 58/30 58 83.2 0.999/91 a the sequences with capital and lowercase letters denote the clamp and core region of the primer, respectively. b Thermal cycling conditions were initially set to 10 min at 95 ○C for the denaturation step, followed by 30 cycles of 95 ○C for 30 s, different annealing temperatures shown in this table for 30 s, a 72 ○C extension for 30 s and a final extension at 72 ○C for 10 min. Final concentration of PCR mixture (25 μl): 0.5U BIOTaq HS DNA polymerase (Bioline, London, UK), 4 mM MgCl2, 0.2 mM dNTP Mix, 0.5 µg·µl-1 bovine serum albumin, 0.2 µM for each primer and 25-50 ng of genomic DNA. In addition, DMSO was used in the PCR amplification of nirK in Cluster III with a final concentration of 5% (v/v) because of the high guanine-cytosine (GC) content of the sequences of Actinobacteria. c The thermal cycling conditions consisted of an initial denaturation step of 98 ○C for 2 min, followed by 40 cycles of 98 ○C for 10 s, different annealing temperatures shown in this table for 10 s and 68○C for 30 s. Final concentration of PCR mixture (20 μl): 10 µl of KOD SYBR qPCR Mix (ToYoBo, Osaka, Japan), 0.4 µl of 50×ROX reference dye, 0.2 µM of primers and 10 ng of environmental DNA. d not detected in the test strains and environmental samples.











