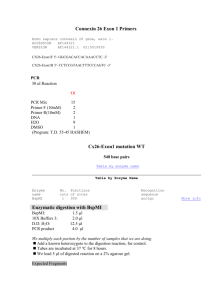
Immunoblot analysis of short-chain enoyl-CoA hydratase

Supplementary material
Synthesis of metabolites
Free acids (crotonic, tiglic, 3(S)-hydroxyisobutyric, 3(R)-hydroxybutyric) were converted to the corresponding acyl chlorides by reaction with thionyl chloride and then separately reacted with an aqueous alkaline solution of glycine, anaqueous alkaline solution of
13
C
2
-glycineor a solution of L-carnitine in trifluoroacetic acid to produce the corresponding acylglycines, acyl
13
C
2
-glycines or acyl-L-carnitines.
Methacrylic and acrylic acids were separately reacted with L-cysteine, 2 H
2
-DL-cysteine
(produced by reduction of
2
H
4
-DL-cystine with tris -(2-carboxyethyl) phosphine), cysteamine or
2
H
4
-cysteamine overnight at pH 7.0 at 37 o
C. Products were isolated by cation exchange chromatography on a column of Dowex 50W-X8 and eluted with 5% ammonia solution.
Fractions containing the expected products were pooled and freeze dried.
Eryththro -2-methyl-2,3-dihydroxybutyrate was prepared by reacting tiglic acid with potassium permanganate.
LC-MSMS metabolite analysis
Urine samples were filtered through 0.2
m cellulose acetate filters and 40
L of the filtrate was mixed with 20
L of internal standard solution containing 10
mol/L of each internal standard listed in Table 1. Samples were dried under an air stream and butylated by heating at
65 o C with 100 µL of n-butanol:acetyl chloride (9:1) for 20 minutes. After decanting and drying down the butylating reagent, samples were reconstituted in 50
L of 50% acetonitrile:water
1
and then diluted with 200
L water. A Waters Acquity TQD system fitted with an
AcquityUPLC BEH C18 1.7 µm 100 x 2.1 mm column. Solvent A was 0.1% acetic acid in water and solvent B was 90% acetonitrile:water containing 0.1% acetic acid. The column was equilibrated with 10% B and 10
L of the sample was injected after which the solvent composition was linearly increased to 60% B at 10 minutes. A flow rate of 0.4 mL/min was used throughout with a column temperature of 30 o C. Multiple reaction monitoring transitions using dwell times of 0.05 sec and conditions given in Supplementary Table 1. Propionyl
2
H
3
carnitine was used as a surrogate internal standard for 3-hydroxyisobutyryl carnitine.
Short-chain enoyl-CoA hydratase enzyme activity measurement
Short-chain enoyl-CoA hydratase (SCEH) activity was measured in fibroblast homogenates using crotonyl-CoA as substrate. Incubations were performed in 100 mM Tris pH 8.0 with
0.005 mg/mL protein at 37°C. Reactions were started with 0.5 mM crotonyl-CoA and stopped after 5 minutes with 2 M HCl to a final concentration of 0.18 M, and subsequently kept on ice for 5 minutes. Samples were neutralised with 2 M KOH plus 0.6 M MES buffer. Methanol
(HPLC grade) was added (final concentration 30% (v/v)). After a further 5 minutes on ice the samples were centrifuged for 5 minutes at 20,000 x g
, 4°C. The supernatant was subjected to ultra-high performance liquid chromatography on a C18 column (Waters Acquity HSS C18 1.8
µm 2.1x100 mm). Resolution of the different CoA-esters was achieved by a linear gradient of methanol (from 18% to 50% (v/v)) in 50 mM potassium phosphate buffer (pH 5.3) at a flow rate of 0.25 mL/min under continuous monitoring at 260 nm. This procedure allowed good resolution of the substrate crotonyl-CoA and the product of the reaction, 3-hydroxybutyryl-
CoA. The amount of 3-hydroxybutyryl-CoA formed was calculated from the ratio of 3hydroxybutyryl-CoA over the total amount of substrate and product, and was used to calculate
2
enzyme activity. This method of quantification corrects for the hydrolysis of the CoA ester by thioesterases present in the homogenate.
Immunoblot analysis of short-chain enoyl-CoA hydratase
Fibroblast homogenates (50 µg) were subjected to electrophoresis on a 10% (w/v) SDSpolyacrylamide gel essentially as previously described (Laemmli, 1970) and transferred to a nitrocellulose sheet. After blocking of non-specific binding sites with 4% normal goat serum in phosphate buffered saline for 1 hour, the blot was incubated overnight at 4 o
C with rabbit polyclonal antibodies raised against bovine liver SCEH (Sigma-Aldrich) diluted 1:1000 in blocking solution. As a loading control, the membranes were re-probed with a monoclonal antibody against beta-actin (Sigma, St Louis, MO, USA) using a 1:10000 dilution (incubation
1 hour). Antigen-antibody complexes were visualized with IRDye 800CW goat anti-rabbit secondary antibody for SCEH and IRDye 680RD donkey anti-mouse secondary antibody for beta-actin using the Odyssey Infrared Imaging System (LI-COR Biosciences, Nebraska, USA).
Goat anti-rabbit IgG antibodies conjugated to alkaline phosphatase, diluted 1:5000 in blocking solution, were used for detection (Santa Cruz Biotechnology, Dallas, TX).
DNA sequencing of
HIBCH and ECHS1 coding exons
Genomic DNA was isolated from skin fibroblasts by standard phenol/chloroform and ethanol precipitation methods. Polymerase chain reaction (PCR)–based methods were used for the molecular analysis. Exons 1 to 14 and flanking intronic regions of HIBCH , and exons 1 to 8 and flanking intronic regions of ECHS1 were amplified using primers designed by the
Exonprimer program. The relevant primers for the mutated ECHS1 exons were: exon 3 forward
3
primer 5'-TATAGGATGGGTGGGCTCTC-3', exon 3 reverse primer 5'-
GTGTATGCACAGCACGGTTC-3', exon 4 forward primer 5'-
AAGCAAGCAGAGTGATTTCTGAC-3' and exon 4 reverse primer 5'-
TGAGACACAGGCAGATTTTGAG-3'. PCR products were subjected to direct sequencing with the same primers used for amplification. The HIBCH and ECHS1 sequences were compared to the reference HIBCH and ECHS1 sequence (Ref Seq NM_014362 and
NM_004092). Two known SNPs were identified in ECHS1 of the first sibling (Exon 2: rs1049951 and Exon 5: rs2230261).
Quantitative PCR of ECHS1 mRNA
Total mRNA was isolated using the TriReagent method (Sigma-Aldrich). The Superscript II
First Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA) was used to generate cDNA. Real time RT-PCR was performed using the ABi7300 Real Time PCR system with
SYBRgreen PCR Master Mix (Applied Biosystems, Foster City, CA). ECHS1 mRNA expression was detected (in triplicate) using the primer set ECHS1 exon 6 forward (5’-
GCGATGGCCAAAGAATCAGTG-3’) and ECHS1 exon 7/8 reverse (5’-
TTCCGGTCATCAGTGGCAAA-3’) (product length 107 bp). Expression was normalised to
ACTB (human beta actin) (forward 5′-AGGCACCAGGGCGTGAT-3′ and reverse 5′-
TCGCCCACATAGGAATCCTT-3′). RNA was isolated from 4 different cell pellets, each was tested at least twice ( n = 9) (each test was performed in triplicate). The results were analysed using the delta-delta comparative C t
method and presented as percent mRNA levels in normal healthy cells.
4
ECHS1 cDNA analysis
ECHS1 cDNA was analysed to determine the effect of the c.414+3G>C splicing mutation (Fig.
2C). PCR of cDNA using the primers exon 3 forward (5’-TGCAGAACCTGAGTTTCCAG-
3’) and exon 4 reverse (5’-ATTAAGATCTCCGGCTGTGC-3’).
5
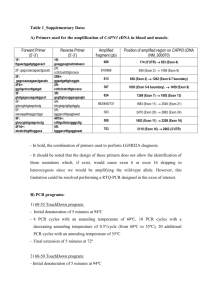
Supplementary Table 1 LC-MSMS characteristics of metabolites as butyl derivatives
Analyte
3-hydroxybutyryl-carnitine
3-hydroxyisobutyryl-carnitine
S-(2-carboxyethyl) 2 H
4
-cysteamine
S-(2-carboxyethyl)cysteamine
Propionyl2 H
3
-carnitine
S-(2-carboxypropyl) 2 H
4
-cysteamine
S-(2-carboxypropyl)cysteamine
Crotonylglycine
Crotonyl 13 C
2
-glycine
S-(2-carboxyethyl)cysteine
S-(2-carboxyethyl) 2 H
2
-cysteine
Tiglylglycine
Tiglyl 13 C
2
-glycine
S-(2-carboxypropyl)cysteine
S-(2-carboxypropyl) 2 H
2
-cysteine
Transition m/z
304.2>85
304.2>85
210.2>105
Cove voltage
35
35
25
206.2>105
277.2>85
224.2>119
220.2>119
200.2>69
202.2>69
306.2>105
25
25
35
25
35
25
25
308.2>105
214.2>83
216.2>83
320.2>119
322.2>119
35
30
30
30
30
25
15
15
20
15
15
25
20
Collision energy
25
25
20
20
25
20
20
Retention time mins
5.58
5.58
6.04
6.04
6.42
6.42
6.63
6.63
2.90/3.08
2.97/3.34
3.14
3.17
3.87
3.92
3.95
6
A
B
C D
Supplementary Figure 1. MRI axial T2 images of sibling 1 at 6 days (A and B) and 2.5 months of age (C and D) showing progressive white matter loss and symmetrical changes in the putamen.
7
O O
H O S
CH
3
NH
2
S-(2-carboxypropyl)cysteine
OH
O
H O S
CH
3
NH
2
S-(2-carboxypropyl)cysteamine
H O
OH
O
O O
H O S
NH
2
S-(2-carboxyethyl)cysteine
OH
O
H O S
NH
2
S-(2-carboxye thyl)cystea mine
H
3
C
CH
3
OH erythro -2-methyl-2,3-dihydroxybutyrate
Supplementary Figure 2 Structures of the metabolites excreted in short-chain enoyl-CoA hydratase deficiency.
8
Mother Father Sibling 1 Sibling 2
Mother
GCTATGCCgt g agtgttgctgccaaga
Father
GCTATGCCgt(c+g)agtgttgctgccaaga
Sibling 1
GCTATGCCgt(c+g)agtgttgctgccaaga
Sibling 2
GCTATGCCgt(c+g)agtgttgctgccaaga
Supplementary Figure 3 Sequencing data for the exon 3 boundary of ECHS1 . The father’s
DNA sequence is the same as his children with a c.414+3G>C mutation affecting the third base of intron 3. The mother does not have this heterozygosity.
9
Supplementary Figure 4 Sequencing of the patient 193bp ECHS1 cDNA (Fig. 2C, lane 1).
Nucleotide position c.473 (arrowed) was predominantly the mutated c.473A allele with a small amount of the wild-type c.473C allele.
10