Supplementary material: Rare familial 16q21 microdeletions under a
advertisement

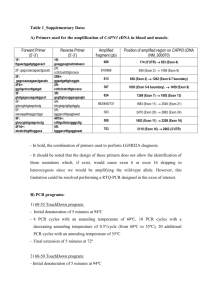
Supplementary material: Rare familial 16q21 microdeletions under a linkage peak implicate cadherin 8 (CDH8) in susceptibility to autism and learning disability Alistair T Pagnamenta1, Hameed Khan2, Susan Walker2, Dianne Gerrelli3, Kirsty Wing1, Maria Clara Bonaglia4, Roberto Giorda5, Tom Berney6, Elisa Mani7, Massimo Molteni7, Dalila Pinto2, Ann Le Couteur6, Joachim Hallmayer8, James S Sutcliffe9, Peter Szatmari10, Andrew D Paterson2, Stephen W Scherer2, Veronica J Vieland11, Anthony P Monaco1 In situ hybridization Human fetal tissue, at 9 weeks of gestation, was dissected and fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) overnight at 4°C. Following fixation, tissues were dehydrated and embedded in paraffin wax. Sagittal sections of the head were cut at 8 µm using a standard microtome and attached to Superfrost Plus microscopic slides (VWR). Before hybridization, tissue sections were de-waxed, hydrated, fixed in 4% PFA/PBS and rinsed twice with PBS. Proteins were removed by incubation with proteinase K (20 mg/ml) in PBS. After washing with PBS, the sections were re-fixed in the same PFA solution, and treated with 0.1 M triethanolamine containing 0.25% acetic anhydride. Slides were dehydrated through an alcohol series and air-dried. Probe regions comprising the 3’-UTR region of the short and long CDH8 isoforms were amplified from whole brain cDNA using primers described in the main text. The resulting amplicon was ligated into pGEM-T (Promega). Antisense and sense probes were prepared by linearising plasmids with SphI and NotI respectively. DigoxigeninUTP was incorporated into riboprobes during in vitro transcription using the DIG RNA labeling mix (Roche) according to the manufacturer’s instructions. Antisense and sense probes were generated using SP6 and T7 polymerase respectively. Hybridization solution contained riboprobe (300ng DIG labeled RNA probe), RNAguard (1 ml/ml) and tRNA (0.5 mg/ml) in hybridization buffer (50% formamide, 0.3 M NaCl, 20 mM Tris-HCl pH 7.5, 5 mM EDTA pH 8.0, 10% dextran sulphate and 1x Denhardt's solution). A 100 µl aliquot of hybridization probe was added to each slide, which was incubated in a sealed chamber moistened with 50% formamide/1x standard saline citrate (SSC) overnight at 65°C. Stringency washes were performed in the following order: 2x SSC (twice at 65°C); 50% formamide/2x SSC (twice at 65°C); 2x SSC (twice at 65°C); 0.2x SSC (65°C) and 0.2x SSC (65°C cooled to room temperature). Slides were then incubated for 1 hour in 150 mM NaCl and 100 mM Tris-HCl pH 7.5 containing 10% fetal calf serum (FCS). For antibody detection, slides were incubated in anti-digoxigenin antibody conjugated with alkaline phosphatase (anti-Dig antibody diluted 1:1000, containing 2% FCS) overnight at 4°C. Expression patterns were visualized using the NBT/BCIP system (Roche). Sections were mounted in VectaMount (Vector Labs) and analyzed using the Axioplan 2 imaging system (Zeiss). Quantitative PCR testing of sample NA18852 Quantitative PCR (qPCR) was used to determine whether a duplication involving CDH8 in HapMap sample NA18852, detected by Wang et al [1] and reported in the Database of Genomic Variants (DGV) [2], is real. qPCR primers were designed for CDH8 coding exons 5-7 and a non-repetitive region of the intron immediately following exon 7, using Primer 3 (Supplementary Table 2 and Supplementary Figure 1). Exon 14 of DOCK4 was used as the control PCR, as genome imbalances in this gene are rare and primers were already available [3]. PCRs were carried out in triplicate in volumes of 25 μl, using 50 ng of DNA, primers at 200 nM and iQ SYBR Green Supermix (BioRad, Hercules, CA), according to the manufacturer’s instructions. Thermocycling and data acquisition was carried out using the iQ5 iCycler (BioRad). Ct outliers were removed in rare cases where the SD of the triplicates was >0.5 cycles. DNA was obtained for HapMap sample NA18852 (Lot B2, 6/7/2006) from Coriell Cell Repositories (Camden, NJ) and we also used family 3099 as these DNA samples had also been derived from cell lines. 3099_009 was chosen as the reference sample, whilst 3099_006 (eldest affected son) was used as a deletion control, as these two samples had similar absorbance profiles to NA18852, as determined by the NanoDrop machine (Thermo Scientific, Waltham, MA). For the five qPCRs reactions, all samples gave mean Ct values between 20.76 and 24.24 and so we deemed it unnecessary to test relative amplification efficiencies. Relative CDH8 dosage was therefore calculated using the 2–ΔΔCt method [4]. We considered that normal copy number results would lie between a threshold of 0.7 and 1.3. The experiment was carried out three times. The whole gene deletion of CDH8 was validated successfully in sample 3099_006. In contrast, the relative CDH8 copy number for sample NA18852 did not increase above 1.3 for any of the CDH8 qPCR probes, in any of the three replicate experiments (Supplementary Figure 1B). There was also no increase between exons 6 and 7 of CDH8, as would be predicted by the CNV call for sample NA18852 listed in the DGV (Supplementary Figure 1). These results are inconsistent with the CDH8 duplication listed for sample NA18852 in the DGV. This duplication was originally detected by only 4 probes from the 550k SNP array in the original study (Supplementary Figure 1A) and was not detected by a subsequent higher-resolution genome-wide CNV scan that used 42 M probes [5]. Therefore we conclude that the CNV call in NA18852 by Wang et al [1] is highly likely to be a false positive result. Supplementary References 1. Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 2007;17(11):1665-74. 2. Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet 2004;36(9):949-51. 3. Maestrini E, Pagnamenta AT, Lamb JA, Bacchelli E, Sykes NH, Sousa I, Toma C, Barnby G, Butler H, Winchester L, Scerri TS, Minopoli F, Reichert J, Cai G, Buxbaum JD, Korvatska O, Schellenberg GD, Dawson G, Bildt AD, Minderaa RB, Mulder EJ, Morris AP, Bailey AJ, Monaco AP. High-density SNP association study and copy number variation analysis of the AUTS1 and AUTS5 loci implicate the IMMP2L-DOCK4 gene region in autism susceptibility. Mol Psychiatry 2009. 4. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001;29(9):e45. 5. Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME. Origins and functional impact of copy number variation in the human genome. Nature 2009;464(7289):704-12. Supplementary Table 1: CDH8 sequencing primers. Exons are numbered starting from the first coding exon that is common to both short and long CDH8 isoforms. * Due to amplicon size, internal primers were also used (available upon request). Sequence Direction GAGCCTATGCTAAGAACCCTCA GGGTCTCACAATCACCCTATTC CGCAAACCAGTTTCTGATAGG CCTTGGGATTAATCGTATTCCT GCAGAAATTGATGATGTTATGTAAAAA ATCCTTGATGCCAGGAGCTA TCCTTTTATTCCCTACACATGGA ATTGATGCAGGCAGTGAAGT TAGTACCAAAATCATGCCATACC TCATTGATGTAACAGGCACACA GGTGCCCAAAAGATATTTGC CACAAGTCCCTCAACTTTAAAACTC GACTCTGAAGTGAAATTTTGATGG TCTTATTGTTCCTGGCTCTGG TCTTCATCAGCATCCATTTTTG TTCCTCTTACTTTAAAATGCAAAGC GGAGAAATGCCCACTGTTTTA ACTCAGCAAATGGTGTTTGAA ACTGATAAGCTGAATTTCAAGTGC GGTAATGCTCTTAATTTTCATCAAC GACGAGTCAGGGAAAAAGCC GGCACTGAAAGTCCTCTTCATTTGG TTTTGTAAAAATACCCCTCTCTCC TTCCTTGCAGGTCCGTATTT Product Size (bp) forward reverse forward reverse forward reverse forward reverse forward reverse forward reverse forward reverse forward reverse forward reverse forward reverse forward reverse forward reverse 1042* Exon amplified Noncoding Region 5'-UTR (long isoform) 5'-UTR and coding 851 Exon 1 478 Exon 2 Coding 344 Exon 3 Coding 480 Exon 4 Coding 343 Exon 5 Coding 781 Exon 6 Coding 688 Exon 7 Coding 1449* Exon 8 Coding 471 Exon 9 Coding 607 Exon 10 Coding 966 Exon 11 Coding and 3'-UTR Supplementary Table 2: qPCR primers, conditions and amplicon details. CDH8 exon 5 CDH8 exon 6 CDH8 exon 7 CDH8 Sequence (5’-3’) AAAAGTGCTGTTCCATCTCCAT CTCAGTACCGGAAGATGTGGTT ATCACGGAGTTTAGAGCAGCAT CAGTCAAAATCGTGGTTGAAGA GAAATGAACAAATCCAGGCACT TAATGCAGACGATGGGAAGATA CCTATGGGGTTCTGCTATTCTG Annealing temperature Amplicon Tm Amplicon size 56°C 80.9°C 123 bp 56°C 81.0°C 100 bp 56°C 80.0°C 117 bp 56°C 76.8°C 111 bp Genome position (NCBI b26) Chr16:6041238360412505 Chr16:6040893260409031 Chr16:6038072660380842 Chr16:60372010- intronic DOCK4 exon 14 TTATTTTCCAAAGCCACAGACC AACCTGTGTGTTCTTCCCTTTG GACCACCTGGGACTGTTGTTAT 59°C 82.5°C 112 bp 60372120 Chr7:111327743111327854 Supplementary Figure 1: Quantitative PCR of the CDH8 gene. A) Customised UCSC plot for chr16:60,350,000-60,420,000 showing qPCR probe positions in relation to the putative duplication #9760 in sample NA18852 (red). B) qPCR results showing CDH8 dosage relative to 3099_009 and results normalised to DOCK4. Mean ±SEM from three experiments are plotted.