MIAPE_Quant_v1.0_SRM_LPCSICUAB_rev
advertisement
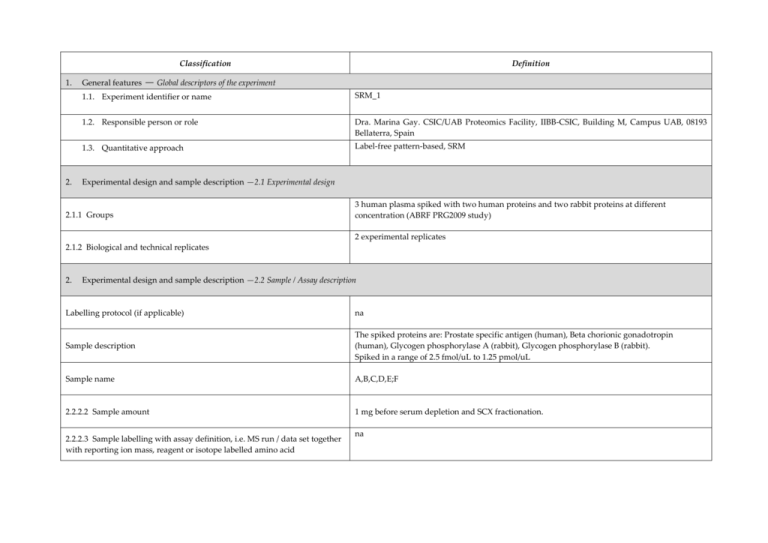
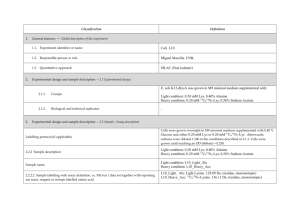
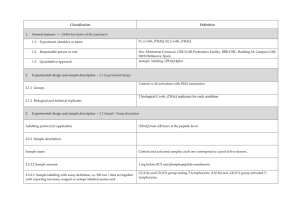
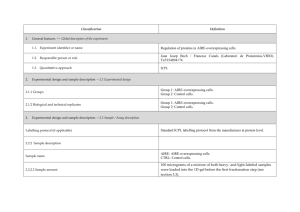
Classification 1. 2. Definition General features — Global descriptors of the experiment 1.1. Experiment identifier or name SRM_1 1.2. Responsible person or role Dra. Marina Gay. CSIC/UAB Proteomics Facility, IIBB-CSIC, Building M, Campus UAB, 08193 Bellaterra, Spain 1.3. Quantitative approach Label-free pattern-based, SRM Experimental design and sample description —2.1 Experimental design 2.1.1 Groups 3 human plasma spiked with two human proteins and two rabbit proteins at different concentration (ABRF PRG2009 study) 2 experimental replicates 2.1.2 Biological and technical replicates 2. Experimental design and sample description —2.2 Sample / Assay description Labelling protocol (if applicable) na Sample description The spiked proteins are: Prostate specific antigen (human), Beta chorionic gonadotropin (human), Glycogen phosphorylase A (rabbit), Glycogen phosphorylase B (rabbit). Spiked in a range of 2.5 fmol/uL to 1.25 pmol/uL Sample name A,B,C,D,E;F 2.2.2.2 Sample amount 1 mg before serum depletion and SCX fractionation. 2.2.2.3 Sample labelling with assay definition, i.e. MS run / data set together with reporting ion mass, reagent or isotope labelled amino acid na 2.2.2.4 Replicates and/or groups A-E, B-F, C-D 2.2.3 Isotopic correction coefficients na 2.2.4 Internal references 1 serum protein (CERU_HUMAN) to normalize the XIC areas 3. 4. Input data — Description and reference of the dataset used for quantitative analysis: type, format and availability of the data. No actual values are requested here. 3.1. Input data type Extracted ion chromatograms (SRM chromatograms) 3.2. Input data format Thermo.raw files 3.3. Input data merging 4 SCX fractions 3.4. Availability of the input data 23 raw files Protocol —Description of the software and methods applied in the quantitative analysis (including transformation functions, aggregation functions and statistical calculations). 4.1. Quantification software name, version and manufacturer Xcalibur 2.1 (ThermoFischer) 4.2. Description of the selection and/or matching method of features, together with the description of the method of the primary extracted quantification values determination for each feature m/z of the precursor and 2 or 3 fragment ions, and retention time for peak selection. For quantitation the area in the extract ion chromatogram was used. and/or peptide 4.3. Confidence filter of features or peptides prior to quantification m/z with 0.5 Da error and retention time with 1 min window. Manual validation. 4.4. Description of data calculation and transformation methods The area of the XIC for the different transitions was used and then normalized with the area obtained from the internal reference. 4.4.1. Missing values imputation and outliers removal na. 4.4.2. Quantification values calculation and / or ratio determination from the primary extracted quantification Average of sample replicates for each condition. values 4.4.3. Replicate aggregation na 4.4.4. Normalization Data normalized using XIC areas of the internal reference Protein quantification values calculation and / or ratio na 4.4.5. determination from the peptide quantification values 4.4.6. 5. Protocol specific corrections na 4.5. Description of methods for (statistical) estimation of correctness PCA analysis to group samples 4.6. Calibration curves of standards na Resulting data —Provide the actual quantification values resulting from your quantification software together with their estimated confidence. Depending of the quantification technique or even of the quantification software, only some of the following items could be satisfied (e.g., for spectral counting, only quantification values at protein level can be provided) 5.1 Quantification values at peptide and/or feature level: Actual quantification values achieved for each peptide and/or, in case of feature-based quantification, for the corresponding features (mapped back from each peptide), together with their estimated confidence. 5.1.1 Primary extracted quantification values for each feature (e.g. area, height, etc.), with their statistical estimation of correctness http://proteo.cnb.csic.es/downloads/miape-quant/Results_peptides_SRM_LPCSICUAB.xlsx 5.1.2 Quantification values for each peptide as a result of the aggregation of the values of the previous section (5.1.1), with their statistical estimation of correctness http://proteo.cnb.csic.es/downloads/miape-quant/Results_peptides_SRM_LPCSICUAB.xlsx 5.2 Quantification values at protein level: Actual quantification values achieved for each protein and for each protein ambiguity group, together with the confidence in the quantification value. Basic / raw quantification values with statistical estimation of correctness http://proteo.cnb.csic.es/downloads/miape-quant/Results_proteins_SRM_LPCSICUAB.xlsx 5.2.2 Transformed / aggregated / combined quantification values of the proteins at group level, with their statistical estimation of correctness http://proteo.cnb.csic.es/downloads/miape-quant/Results_proteins_SRM_LPCSICUAB.xlsx