Proteome-Dicoverer-key-words-and
advertisement

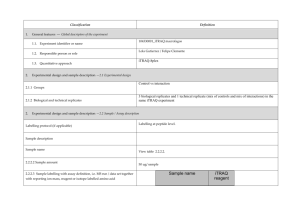
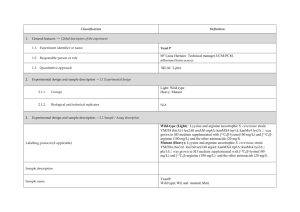
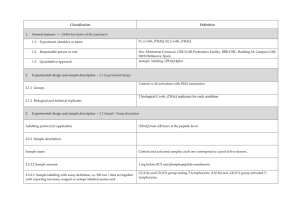
PROTEOME DISCOVERER- software for determination of Relative Quantitation (Thermo Scientific) explained ver 1/23/2013 Quan Info (in .msf files with quantification results only) indicates why a peptide was used in quantification or why it was not: • No Quan Values: No isotopic or isobaric labels were detected in the sample. i.e., reductive methylation labeling failure n sample(s) • No Quan Labels: The peptide was not modified by a label (no light, medium or heavy). I.e., reductive methylation label failure of that peptide- check your .xls sheet to see the labeling efficiency of this experiment. Normally our lab is able to achieve 97% or better in labelling of the peptides • Redundant: The same MS/MS spectra were used for two separate peptide matches. • No Proteins: The peptide is not associated with a protein, probably because it did not go above the cut-off threshold defined by the search engines, using these parameters: the Protein Cutoff Score parameter in Mascot, the Protein XCorr parameter in SEQUEST, or the Protein Score Cutoff parameter in ZCore, i.e., too ambiguous to be IDd • Unique: The same MS/MS spectra were used for one peptide match, and the peptide belongs to one protein. • Not Unique: The SAME peptide was found in another protein. • Indistinguishable Channels: The peptide does not have amino acids that could have the specified labels or it has the specified labels but has defined channels with the same delta masses, rendering the channels indistinguishable, i.e., can’t tell if the peptide is from light, medium or heavy labeled sample • Excluded by Method: Peptides or quantification results are excluded from protein quantification according to the Single-Peak/Missing Channels Allowed setting in the quantification method. This setting specifies the allowed number of missing quantification channels or single-peak quantification channels that can be included in valid quantification results in the ratio calculation. • Inconsistently Labeled: Peptides do not always display the expected amount of isotopic labeling. • Filtered Out: Peptides are not used in the quantification because the applied filters removed them. This column appears only when you have loaded an .msf file containing quantification results. Tutorial time: Relative Quantitation with Proteome Discoverer The H/L and M/L ratios reflect the area under the peak in the MS spectrum for that specific peptide which contributes to the ID of a protein. The ratios then would compare the peak area of that signal intensity or basically the amount of that peptide So if you have in the protein view on the .xls, a ratio in bold blue of 4.73 for M/L then that would mean the medium labeled sample had 4.43/1 or 4.73 TIMES more of that protein than the light labeled sample If you had a bold pink number of say .255 in the H/L that would mean you had 1/0.255 or 3.92 X more of that protein in the (light) than the (Heavy) Looking at the excel spreadsheet we sent you- on your computer you can click on a + on the far left of the protein .xls to expand and see the associated peptides that contributed to that protein ID as well as the ratio (or not) of the M/L or H/L. There you can see how many of the labels are L, M or H labeled. I f you click on the #2 at the top far left of the spreadsheet you can expand all the proteins peptide lists for the entire sheet. When looking for the proteins that are *missing* in any of the samples you would have to 1st look for proteins w/o any ratio calculated, 2nd-then, expand that protein as described above with the + and insure the ID is based on more than 2 peptides, 3rd - check to see if there is all L or M or H peptides - if there are 8 H peptides and 0 light- then this would indicate the heavy sample had this protein and the Light sample did not. Same thing would be true of MASCOT- just look down the list to see what is ID’d and look at the labels for the peptides that contribute to that ID and what sample they came from. Relative Quantitation cannot be calculated in the case of the protein not being present in one sample and is present in another!