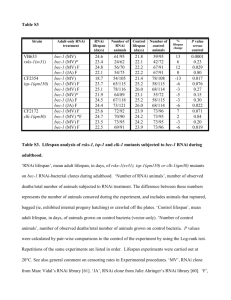
Background for the upcoming discussion paper In the late 1990`s
advertisement

Background for the upcoming discussion paper In the late 1990’s/early 2000’s Andy Fire and Craig Mello and members of their two labs had found that double stranded RNA triggers gene silencing through a post-transcriptional processes that they named RNA interference (RNAi). To better understand this process, the Mello lab searched for mutants that exhibited defects in RNAi. One set of such mutants were named rde (RNA interference defective). In setting up the genetic screen to identify these rde mutants, the Mello lab took advantage of several aspects of worm biology. They knew that loss of function mutations in a gene called pos-1 (see below for more detail) causes a maternal effect embryonic lethal phenotype (go back to your class notes to review maternal effect). In other words, pos-1 is an essential gene for embryonic development. Hermaphrodites exposed to feeding pos-1 RNAi are unaffected but produce dead embryos. Mutant animals resistant to pos-1 RNAi produce viable progeny. To facilitate identification of mutants, they used a starting strain that is vulvaless [lin-2(e1309), which gives the “bag of worms” phenotype]. When lin2(e1309) mutants are exposed to pos-1 RNAi, the embryos all die and since the animals cannot lay their eggs anyway, all of the dead embryos will be contained within the hermaphrodite parent. To find rde mutants, they screened for animals that could now produce viable progeny even when exposed to pos-1 RNAi. The discussion paper investigates whether the rde mutations and another set of mutations also found that cause defects in RNAi play any roles in transmission of RNAi from one generation to another. The authors investigate to consequences of RNAi mediated inactivation of each of three genes. In the context of this paper, these genes are only relevant in that they cause embryonic lethal phenotypes, why they do so is not really pertinent here. pos-1 (posterior segregation mutant) encodes a CCCH-type zinc-finger protein; during embryogenesis, maternally provided POS-1 is essential for proper fate specification of germ cells, intestine, pharynx, and hypodermis; POS-1's role in cell fate specification is likely as a translational regulator. mom-2 encodes a member of the Wnt family of secreted signaling glycoproteins that is required for induction of endodermal (gut) tissue in the 4-cell stage embryo; MOM-2, required in the inducing blastomere (P2) at the 4-cell stage. sgg-1=gsk-3 encodes the C. elegans glycogen synthase kinase ortholog; during embryonic development, GSK-3 functions in the Wnt signaling pathway that restricts specification of mesendodermal tissue to the appropriate blastomere. mDf-3: a deficiency that corresponds to the chromosomal region of pos-1. The key point of the discussion will be deciphering the logic of the crosses in each of the figures. Why did they use the specific genetic backgrounds indicated? How else might you go about addressing the questions that are addressed in this paper?