Table S1. RNAi phenotypes of lsm genes retrieved from wormmart
advertisement

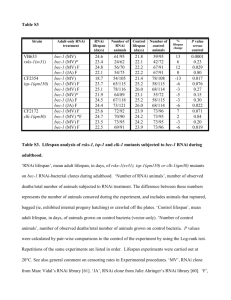
Table S1. RNAi phenotypes of lsm genes retrieved from wormmart. Wormmart is a tool available at wormbase [S1]. RNAi assays represented in this table were performed following diverse protocols and using distinct genetic backgrounds: RNAi by feeding starting at L4 [S2], RNAi by feeding starting at L1 [S3], RNAi in RNAi hypersensitive backgrounds [S4, S5], RNAi by injection [S6], or RNAi by soaking [S7]. Other RNAi screens referred here were focused in specific phenotypes as aging, cell proliferation, endocytosis, innate immunity or transgene silencing [S8–13]. Table S2. Yeast lsm phenome. Table shows all the yeast lsm phenotypes retrieved from the Saccharomyces Genome Database (SGD) [S13]. All the information available for S. cerevisiae mutant strains for lsm genes is shown. Nonviable phenotypes are highlighted in red. Table S3. RNA-Seq analyses of lsm-1 mutants. Sheet 1: Statistical analyses including FPKM (fragments per kilobase of exon per million fragments mapped) for each gene mapped, and fold change between lsm-1 mutants and wild type animals. Sheet 2: Genes significantly upregulated (p value ≤ 0.05). Sheet 3: Genes significantly downregulated (p value ≤ 0.05). Sheets 4 and 5: Additional information about genes up and down regulated in lsm-1 mutants. Gene description was retrieved from wormbase using the tool wormmart. References for Supplementary Data S1. Schwarz EM et al. (2006) WormBase: better software, richer content. Nucleic Acids Res 34:D475–8. S2. Kamath RS et al. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421:231–7. S3. Rual J-F et al. (2004) Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res 14:2162–8. S4. Simmer F et al. (2003) Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol 1:E12. S5. Ceron J et al. (2007) Large-scale RNAi screens identify novel genes that interact with the C. elegans retinoblastoma pathway as well as splicing-related components with synMuv B activity. BMC Dev Biol 7:30. S6. Sönnichsen B et al. (2005) Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434:462–9. S7. Maeda I, Kohara Y, Yamamoto M, Sugimoto A (2001) Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol 11:171–176. S8. Robert VJP, Sijen T, van Wolfswinkel J, Plasterk RHA (2005) Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev 19:782–7. S9. Samuelson A V, Carr CE, Ruvkun G (2007) Gene activities that mediate increased life span of C. elegans insulin-like signaling mutants. Genes Dev 21:2976–94. S10. Balklava Z, Pant S, Fares H, Grant BD (2007) Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat Cell Biol 9:1066–73. S11. Hebeisen M, Drysdale J, Roy R (2008) Suppressors of the cdc-25.1(gf)- associated intestinal hyperplasia reveal important maternal roles for prp-8 and a subset of splicing factors in C. elegans. RNA 14:2618–33. S12. Alper S et al. (2008) Identification of innate immunity genes and pathways using a comparative genomics approach. Proc Natl Acad Sci U S A 105:7016–21. S13. Cherry JM et al. (2012) Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40:D700–5.