tpj12151-sup-0016
advertisement

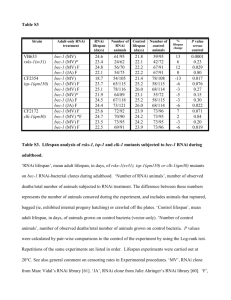
1 Legends for Supplementary Material 2 3 Figure S1. Comparison of Brassica TT16 genomic sequences. Exons and introns were 4 labelled with black and white backgrounds respectively. Bn, B. napus; AA, B. rapa; CC, 5 B. oleracea. The TT16 genomic DNA sequences cloned in this study were submitted to 6 GenBank as follows: BnTT16.1, JF970611; BnTT16.2, JF970612; BnTT16.3, JF970613; 7 BnTT16.4, JF970614; BoTT16.1, JF970615; BoTT16.2, JF970616; BoTT16.3, JF970617; 8 BoTT16.4, JF970618; BrTT16.1, JF970619; BrTT16.3, JF970620; BrTT16.4, JF970621. 9 10 Figure S2. Schematic diagram of BnTT16 overexpression constructs used to complement 11 the Arabidopsis tt16-6 mutant phenotype. 12 13 Figure S3. Localization of proanthocyanidins using p-dimethylaminocinnamaldehyde 14 staining in developing seeds of complemented Arabidopsis lines. (a) wild-type 15 Arabidopsis Ws-3 as a positive control; (b) Arabidopsis transparent testa 16-6 (Attt16-6) 16 mutant as a negative control; (c) representative complemented line expressing Brassica 17 napus BnTT16.1 (Attt16-6/35S::BnTT16.1); (d) representative complemented line 18 expressing BnTT16.2 (Attt16-6/35S::BnTT16.2); (e) representative complemented line 19 expressing BnTT16.3 (Attt16-6/35S::BnTT16.3); (f) representative complemented line 20 expressing BnTT16.4 (Attt16-6/35S::BnTT16.4). Scale bar = 100 µm. The arrows indicate 21 the endothelium containing PA. 22 1 23 Figure S4. RNAi-mediated silencing of BnTT16 expression. (a) Schematic diagram of 24 the construct utilized for RNAi silencing of BnTT16 homologs in B. napus. (b) The 25 overall level of expression of BnTT16 genes in open flowers (0-DAP) was down- 26 regulated in Bntt16 RNAi lines. (c) The expression level of B. napus AGAMOUS-LIKE 6 27 (BnAGL6) exhibited no significant difference between wild-type (WT) and Bntt16 RNAi 28 lines (RNAi) (p < 0.05). The data in (b) and (c) were presented as the relative expression 29 levels (2-ΔCT) of target genes compared with reference genes (Table S1). 4-1, 10-3, 12-4, 30 and 23-4 are 4 representative Bntt16 RNAi lines. 31 32 Figure S5. Transcriptional expression (2-ΔCT) of each individual BnTT16 gene in open 33 flowers (0 day after pollination) was down-regulated in RNAi plants. (a) BnTT16.1; (b) 34 BnTT16.2; (c) BnTT16.3; (d) BnTT16.4. 4-1, 10-3, 12-4, and 23-4 are 4 representative 35 Bntt16 RNAi lines. The data were presented as the relative expression levels of each 36 BnTT16 gene compared with reference genes listed in Table S1. 37 38 Figure S6. Vanillin staining of whole developing seeds (14 day after pollination) 39 indicated that proanthocyanidin content was less in Bntt16 RNAi plants than wild-type 40 plants. (a) Wild-type whole seeds; (b) halved wild-type seeds; (c) whole seeds from 41 RNAi plants; (d) halved seeds from RNAi plants. Bars = 1 mm. 42 43 Figure S7. Localization of proanthocyanidins with vanillin in developing seeds of B. 44 napus wild-type (WT) and RNAi lines. Arrows indicate PAs that have accumulated in the 45 endothelium. (a) WT 15-DAP; (b) RNAi 15-DAP; (c) WT 20-DAP; (d) RNAi 20-DAP; 2 46 (e) WT 24-DAP; (f) RNAi 24-DAP; (g) WT 31-DAP; (h) RNAi 31-DAP. RNAi, Bntt16 47 RNAi plants; DAP, days after pollination. Scale bar = 100 µm. 48 49 Figure S8. Global similarity of the eight microarray samples for wild-type B. napus and 50 RNAi plants. 51 Sample clustering was used to assess the overall similarity of the eight samples. The four 52 replicates of wild-type plants (sample 1-4) clustered with one another, while the four 53 replicates of RNAi plants formed a separate group (sample 5-8). 54 55 Figure S9. Validation of microarray data (fold-difference) using qRT-PCR. (a) Candidate 56 genes involved in flavonoid biosynthesis and regulation. (b) Candidate genes involved in 57 seed coat development. Data include the mean relative transcript levels ± standard error 58 of four biological replicates and four technical replicates. 59 60 Figure S10. Effects of Bntt16 silencing on the gene expression profile of B. napus (n=4). 61 (a) Biological process. (b) Molecular function. (c) Cellular component. GO, gene 62 ontology. Unknown and ambiguous genetic programming categories were not included. 63 64 Figure S11. Vanillin staining of representative dissected developing seeds. Samples were 65 harvested at 15 day after pollination (15-DAP, cotyledon stage) from wild-type B. napus 66 plants. (a) embryo; (b) endosperm; (c) inner integument; (d) epidermis. Bar = 1 mm. 67 Briefly, we dissected 15-DAP developing seed coats into three contiguous components: 68 inner fraction composed primarily of developing endosperm, middle fraction containing 3 69 inner integument and palisade cells, and outer fraction composed of epidermal and sub- 70 epidermal cells (Jiang and Deyholos 2010). To detect which fraction contained the 71 endothelium, we stained these three fractions and embryos with vanillin. The middle 72 fraction highly stained to red, and the inner fraction was only slightly stained. Since 73 almost all PA accumulated in the endothelium at 15-DAP (Figure 7), we confirmed that 74 the middle fraction contained virtually all the endothelium. For convenience, we named 75 the four sub-tissue fractions as embryo, endosperm, inner integument and epidermis in 76 this study. 77 78 Table S1. Primers utilized in this study. 79 80 Table S2. Microarray data 81 82 Table S3. Proteins used for phylogenetic analysis. 4