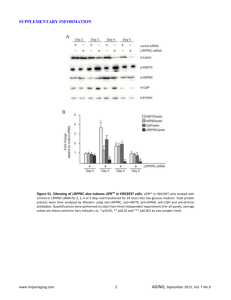
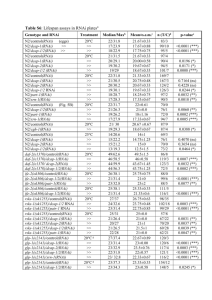
tpj12302-sup-0001-AppendixS1
advertisement

Appendix 1 Supporting Methods Virus-induced gene silencing (VIGS) The NbPES cDNA fragments were PCR-amplified and cloned into the pTV00 vector containing part of the tobacco rattle virus (TRV) genome using the NbPES(F)-F/R, NbPES(N)-F/R, and NbPES(C)-F/R primer sets as described in Supplementary Table S2. For VIGS of NbBOP1 and NbWDR12, the corresponding cDNA fragments were PCR-amplified with the NbBOP1-F/R and NbWDR12-F/R primer sets. VIGS was performed as described (Ahn et al., 2011). Agrobacterium-mediated transient expression Agro-infiltration was carried out as described (Voinnet et al., 2003). Agrobacterial cultures (GV3101) containing various constructs fused to the CaMV35S promoter were adjusted to OD600=0.6 in MES buffer (10 mM MES [pH 7.5], 10 mM MgSO4). The suspension was incubated with acetosyringone for 2–3 h at a final concentration of 150 μM, and infiltrated into leaves of wild-type N. benthamiana plants. In all experiments, Agrobacterium C58C1 carrying the 35S:p19 construct (Voinnet et al., 2003) was coinfiltrated to achieve maximum levels of protein expression. Expressed proteins were analyzed at 48 h post-infiltration. Generation of dexamethasone (DEX)-inducible AtPES RNAi lines in Arabidopsis 1 A 350 bp AtPES cDNA fragment was PCR amplified using the AtPESi-S-F/R primer set containing XhoI and ClaI sites for the sense construct, and the AtPESi-AS-F/R primer set containing BamHI and SpeI sites for the antisense construct. The sense and antisense constructs of the 350 bp cDNA regions were cloned into the 5' and 3' arms of the intron in the pSK-int vector (Guo et al., 2003) to produce an inverted-repeat sequence (antisense-intron-sense), which was isolated by digestion with XhoI and SpeI, and cloned into the binary vector pTA7002 (Aoyama et al., 1997). The recombinant plasmid containing the AtPES RNAi construct was introduced into Agrobacterium tumefaciens (C58C1), and transformed into Arabidopsis (Col-0 ecotype) using Agrobacterium-mediated transformation. Transgenic plants were selected on growth media containing hygromycin (30 mg/L). For induction of RNAi, dexamethasone (Sigma) was added to the medium to a final concentration of 10 μM in ethanol (0.033%) and tween 20 (0.01 % w/v) from 30 mM stock solution. Generation of DEX-inducible NbPES RNAi BY-2 cell lines A 300 bp NbPES cDNA fragment was PCR amplified using the BY2 PESi-S-F/R primer set containing XhoI and ClaI sites for the sense construct, and BY2 PESi-AS-F/R primer set containing BamHI and SpeI sites for the antisense construct. The sense and antisense constructs of the 300 bp cDNA regions were cloned into the 5' and 3' arms of the intron in the pSK-int vector (Guo et al., 2003) to produce an inverted-repeat sequence (antisense-intron-sense), which was isolated by digestion with XhoI and SpeI, and cloned into pTA7002 vector (Aoyama et al., 1997). 2 The recombinant plasmid containing the NbPES RNAi construct was introduced into Agrobacterium tumefaciens (GV3101), and transformed into tobacco BY-2 cells using Agrobacterium-mediated transformation (Nakashima et al., 2000). Transgenic BY-2 cells were selected on media containing hygromycin (50 mg/L) and maintained at 25°C in the dark with weekly subculture in modified Linsmaier and Skoog medium (Banno et al., 1993) containing hygromycin (50 mg/L). For induction of RNAi, dexamethasone (Sigma) was added to the medium to a final concentration of 10 μM from 30 mM stock solution. Bimolecular fluorescence complementation (BiFC) BiFC analyses were performed as described by Ahn et al. (2011). The coding region of AtPES, BOP1, and WDR12 was PCR-amplified and cloned into the pSPYNE vector containing the N-terminal region of YFP (amino acid residues 1-155). Similarly, AtPES, BOP1, and WDR12 cDNAs were cloned into pSPYCE vector containing the Cterminal region of YFP (residues 156–239). The pSPYNE and pSPYCE fusion constructs were agroinfiltrated together into the leaves of 3-week-old N. benthamiana plants as described (Walter et al., 2004). After 48 h, protoplasts were generated and YFP signal was detected using a confocal laser scanning microsope (Zeiss LSM510). Real-time quantitative RT-PCR Real-time quantitative RT-PCR was carried out using the primers in Supplementary Table S2 as described (Lee et al., 2009). For detection of rRNA precursors containing ITS1 and ITS2 sequences, real-time quantitative RT-PCR was performed with ITS1 (5’- 3 ggtttcgcgtatcggcatg-3’ and 5’-cgtgccggggttttgtgata-3’) and ITS2 (5’- catgcggtggtgaacttgat-3’ and 5’-tgggtcatctacagcttccg-3’) primer sets. DAB staining H2O2 accumulation in plant tissues was detected by 3,3’-diaminobenzidine (DAB) staining as described (Thordal-Christensen et al., 1997). Leaves of Arabidopsis seedlings were incubated in 1 mg/mL 3,3’-diaminobenzidine Tetrahydrochloride [pH 3.8] (Sigma) for 8 h at room temperature. Then the leaves were cleared by boiling in 96% ethanol for 10 min. H2O2 is visualized as a reddish-brown coloration. Deoxynucleotidyl transferase-mediated dUTP nick end labelling (TUNEL) assay Leaves from AtPES RNAi Arabidopsis plants were fixed and treated with DNase-free RNase A (30 μg/mL; Promega) and Proteinase K (20 μg/mL; Qiagen) for 5 min before TUNEL assay. TUNEL assay was carried out according the manufacturer’s manual (Promega), and counterstained with DAPI solution (0.1 μg/mL DAPI, 50 mM NaCl, 5 mM EDTA, 10 mM mercaptoethylamine, and 10 mM Tris-HCl [pH 7.4]) for 5-10 min before examination with confocal microscopy (Zeiss LSM510). For a positive control, leaves from the ethanol-treated RNAi plants (-DEX) were treated with RQ1 RNase-Free DNase (Promega) for 15 min at 37oC before TUNEL assay according the manual (Promega). Transmission electron microscopy 4 Tissue sectioning and transmission electron microscopy were carried out using the fourth or fifth leaf above the infiltrated leaf of the VIGS lines as described by Ahn et al. (2004). Co-immunopreciptation Co-immunopreciptation was performed as described (Ahn et al., 2011). Flag-tagged AtPES and BOP1, HA-tagged WDR12, and Myc-tagged BOP1 were co-expressed in combination in N. benthamiana leaves by agroinfiltration. After 48-h incubation, leaf extracts were prepared in 300 μL immunoprecipitation buffer (25 mM Tris [pH 7.5], 150 mM NaCl, 1 mM DTT, 1 mM CaCl2, 5 mM NaF, 25 mM Na3VO4, 1 mM PMSF, 25 mM glycerophosphate, protease inhibitor cocktail, 0.1 % Triton X-100, and 0.5 % NP40). After brief centrifugation to remove cell debris, protein extracts (1 mg) were incubated with 1 μg of monoclonal mouse anti-HA antibody (ABM; IP grade) or 1 μg of monoclonal mouse anti-Myc antibody (ABM; IP grade) at 4°C for overnight, and then 25 μL packed volume of Protein A-agarose resin (Pharmacia) was added to the mixture for further incubation at 4°C for 4 h. After brief centrifugation, the resin was washed 3 times with the immunoprecipitation buffer, and then resuspended in 2X SDS sample buffer. After boiling, the resin was precipitated by brief centrifugation, and the supernatant was run on 10% SDS-PAGE. Western blotting was carried out using the monoclonal anti-Flag antibody (ABM; 1:10,000), monoclonal anti-Myc antibody (ABM; 1:10,000), and monoclonal anti-HA antibody (ABM; 1:10,000). 5 Incorporation of 35S-labelled methionine 7 day-old Arabidopsis seedlings of AtPES RNAi lines were transferred to media with ethanol or DEX (10 μM) for further growth for 4 days. Using 35S-labelled methionine, newly synthesized proteins were detected as described (Lageix et al., 2008). Sucrose gradient sedimentation Sucrose gradient sedimentation was performed as described (Kwon and Cho, 2008). 0.2 g of fine frozen plant tissue powder were homogenized in 1 ml PEB buffer (40 mM Tris–HCl [pH 8.0], 20 mM KCl, 10 mM MgCl2, 100 mg/ml heparin, 100 μg/ml chloramphenicol, 25 μg/ml cycloheximide). Cell debris was removed by centrifugation at 16,000 g for 15 min at 4 oC, and 0.5 ml of the supernatant was loaded onto a 4.4 ml 10-50 % sucrose cushion in PEB buffer, and then centrifuged at 45,000 rpm for 65 min at 4 oC using a SW 55Ti rotor (Beckman). Fractions of 400 μl each were collected and 30 μl of each selected fraction was analyzed by SDS-PAGE and immunoblotting. Immunoblotting was performed according to the manufacturer’s instructions using monoclonal mouse antibodies against the HA tag (1:10,000 dilution; ABM), the Flag tag (1:10,000; ABM), and the 60S ribosomal protein L10a (1:10,000; Santa Cruz), and then with horseradish peroxidase-conjugated goat anti-mouse IgG antibodies (Promega) and ECL chemiluminescence kit for detection (GE Healthcare Life Sciences). Metabolic labeling of rRNA 6 Metabolic labeling of rRNA was performed according to Lange et al. (2011). Equal fresh weights (100 mg each) of wild-type and AtPES RNAi seedlings were incubated for 4 h with 500 μCi [α-32P]-UTP that was diluted in 1 ml sterile water. Total RNA was extracted with a SpectrumTM Plant Total RNA kit (Sigma), according to manufacturer instructions. Uptake of the radioactive UTP was estimated by scintillation counting of crude extracts. RNA samples, separated by agarose gel electrophoresis, were blotted on Hybond NX membranes (Amersham) and analyzed with a phosphorimager (Molecular Dynamics). Ribosome profiling Ribosomes were isolated from N. benthamiana leaves as described by Ahn et al. (2011). Frozen tissues were homogenized in 2 mL PEB (200 mM Tris-HCl (pH 9.0), 200 mM KCl, 25 mM EGTA, 36 mM MgCl2, 5 mM dithiothreitol, 50 mg/mL cycloheximide, 50 mg/mL chloramphenicol, 0.5 mg/mL heparin, 1% Triton X-100, 1% Tween 20, 1% Brij-35, 1% deoxycholic acid, 1% Igepal CA-630, and 2% polyoxyethylene) per mL of tissue. Cell debris was removed by centrifugation at 16,000 g for 15 min at 4°C, and the supernatant was loaded onto an 8-mL 1.6 M sucrose cushion and centrifuged at 170,000 g for 18 h. Pellets were resuspended in 700 μL R buffer (200 mM Tris-HCl [pH 9.0], 200 mM KCl, 25 mM EGTA, 36 mM MgCl2, 5 mM DTT, 50 mg/mL cycloheximide, and 50 mg/mL chloramphenicol) and incubated at 4°C for 1 h, as described by Zanetti et al. (2005). The ribosomes were layered onto an 11-mL 15%–60% sucrose gradient and centrifuged at 170,000 g for 3 h at 4°C, and the light absorbance of each fraction was measured at 254 nm with a fraction collection system. 7 Immunofluorescence Preparation of BY-2 cells and immunofluorescence were carried out as described by Lee et al. (2009). For double-labeling of Flag and α-tubulin, fixed and permeabilized BY-2 cells that express NbPES:Flag fusion proteins were immunolabeled with a 1:500 dilution of anti-Flag (IgG3) antibodies (Applied Biological Materials) and a 1:1000 dilution of anti-α-tubulin (IgG2a) antibodies (DM1A; Sigma). Next, the cells were incubated with a 1:1000 dilution of IgG3-specific Alexa Fluor 593-conjugated antimouse IgG antibodies (Molecular Probes); and a 1:1000 dilution of IgG2a-specific Alexa Fluor 488-conjugated anti-mouse IgG antibodies (Molecular Probes). Finally, cells were stained with 0.2 µg/mL DAPI and observed under a confocal laser scanning microscope (Carl Zeiss LSM 510). Statistical Analyses Two-tailed Student’s t-tests were performed using the Minitab 16 program (Minitab Inc.; http://www.minitab.com/en-KR/default.aspx) to investigate the statistical differences between the responses of the samples. Significant differences between control and other samples were indicated by one (P ≤ 0.05) or two (P ≤0.01) asterisks. 8 SUPPORTING REFERENCES Ahn, C.S., Han, J.A., Lee, H.S., Lee, S. and Pai, H.S. (2011) The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell 23, 185-209. Ahn, J.W., Kim, M., Kim, G.T., Lim, J.H. and Pai, H.S. (2004) Phytocalpain controls the proliferation and differentiation fates of cells in plant organ development. Plant J. 38, 969–981. Aoyama, T. and Chua, N.-H. (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11, 605-612. Banno, H., Hirano, K., Nakamura, T., Irie, K., Nomoto, S., Matsumoto, K. and Machida, Y. (1993) NPK1, a tobacco gene that encodes a protein with a domain homologous to yeast BCK1, STE11, and Byr2 protein kinases. Mol. Cell Biol. 13, 4745–4752. Guo, H.-S., Fei, J-F., Xie, Q. and Chua, N-H. (2003) A chemical-regulated inducible RNAi system in plants. Plant J. 34, 383 –392. Kwon, K.C. and Cho, M.H. (2008) Deletion of the chloroplast-localized AtTerC gene product in Arabidopsis thaliana leads to loss of the thylakoid membrane and to seedling lethality. Plant J. 55, 428-442. Lageix, S., Lanet, E., Pouch-Pélissier, M.N., Espagnol, M.C., Robaglia, C., Deragon, J.M. and Pélissier, T. (2008) Arabidopsis eIF2α kinase GCN2 is essential for growth in stress conditions and is activated by wounding. BMC Plant Biol. 8, 134. 9 Lee, J.Y., Lee, H.S. Wi, S.J. Park, K.Y., Schmit, A.C. and Pai, H.S. (2009) Dual functions of Nicotiana benthamiana Rae1 in interphase and mitosis. Plant J. 59, 278–291. Nakashima, K., Shinwari, Z. K., Sakuma, Y., Seki, M., Miura, S., Shinozaki, K. and Yamaguchi-Shinozaki, K. (2000) Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydrationand high-salinity-responsive gene expression . Plant Mol. Biol. 42, 657–665. Thordal-Christensen, H., Zhang, Z.G., Wei, Y.D. and Collinge, D.B. (1997) Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley- powdery mildew interaction. Plant J. 11, 1187–1194. Voinnet, O., Rivas, S., Mestre, P. and Baulcombe, D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956. Walter, M., Chaban, C., Schütze, K., Batistic, O., Weckermann, K., Näke, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K. and Kudla, J. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428-438. Zanetti, M.E., Chang, I.F., Gong, F., Galbraith, D.W. and Bailey-Serres, J. (2005) Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol. 138, 624–635. 10