nph12217-sup-0001-FigS1-S10_TableS1
advertisement

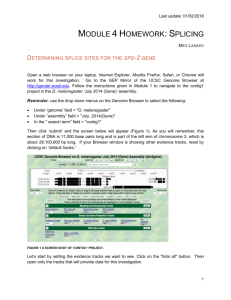
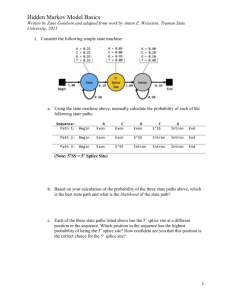
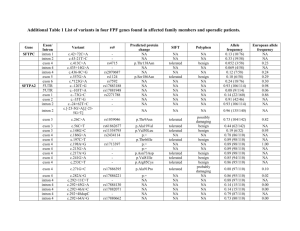
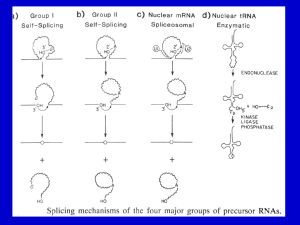
Supporting Information Figs S1–S10 and Table S1 Fig. S1 The 530 bp sequence of the StCCD8 gene introduced into the pHellsgate8 vector, to generate the StCCD8-RNAi construct Fig. S2 Alignment of the known and putative CCD8 proteins sequences; Ph, Petunia hybrida; Sl, Solanum lycopersicum; St, Solanum tuberosum, using the Jalview software (http://www.jalview.org/). Fig. S3 Formation of ‘aerial’ tubers on the lower nodes of the stems of StCCD8-RNAi (line 1) potato plants, with new shoots developing from the apical and lateral tuber buds. Fig. S4 Phenotype of DM tubers grown in vitro from node stem cuttings. Fig. S5 Distribution of number (a) and yield (b) of potato tubers in StCCD8-RNAi potato lines compared to controls- wild type (potato cv Desiree) and EV_CCD8 potato. Values are based on analysis of 60-d-old plants grown from tissue culture stem cuttings, where n=14. Error bars indicate ± SE (standard error). Statistical differences from the wild type control were calculated with Student’s one-tailed t test, assuming unequal variance. An asterisk was added where there was a significant difference between the calculated (P <0.05) values for RNAi lines and controls. Fig. S6 Sprouting of StCCD8-RNAi potato (line 1), compared to potato cv Desiree, at 3 months after harvest. Fig. S7 Chlorophyll content in StCCD8-RNAi plants. (a) Comparison of leaf colour from transgenic and control potato cv Desiree plants. (b) Total chlorophyll content in leaf tissue of Desiree and RNAi lines of potato plants. Samples were obtained from leaf discs taken from the 3rd node of the main stem of 60 d old plants grown from tubers. Scale bar= 5 cm. Values are means of triplicate assays ± SE (standard error), using tissue sampled from 4 independent biological replicates. Statistical differences from the wild type control were calculated with Student’s one-tailed t test, assuming unequal variance. An asterisk was added where there was a significant difference between the calculated (P <0.05) values for RNAi lines and controls. Fig. S8 Total carotenoid content in tuber and root tissue of StCCD8-RNAi potato plants, in the first season of growth. Values are based on analysis of 60 (developing) and 90 (mature) d old plants grown from tissue culture stem cuttings, where n=3. Error bars indicate ± SE (standard error). Statistical differences from the wild type control were calculated with Student’s one-tailed t test, assuming unequal variance. An asterisk was added where there was a significant difference between the calculated (P <0.05) values for RNAi lines and controls. Fig. S9 Comparison of the promoter and exon/intron structure for the StCCD8 gene (A) published sequence data available from the potato genome browser and (B) generated data from Desiree. The grey boxes represent areas of inserted sequence and the black boxes the CCD8 exons. CCD8 coding sequence in DM and Desiree is 1674bp = 558 amino acids. DM CCD8 sequence: 3802 nt in total, exon 1-297 nt, exon 2-744 nt, exon 3-195 nt, exon 4-189 nt, exon 5-105 nt, exon 6-144 nt, intron 1-1189 nt, intron 2-611, intron 3-112, intron 4-72 nt, intron 5-145 nt. Desiree CCD8 sequence: 3462 nt in total, exon 1-297 nt, exon 2-747 nt, exon 3-189 nt, exon 4-192 nt, exon 5-105 nt, exon 6-144 nt, intron 1-860 nt, intron 2-589, intron 3-119, intron 4-75 nt, intron 5-145 nt Insertion sizes: 1= 48bp, 2= 138bp, 3= 336bp, 4= 18bp, 5= 21bp, 6= 13bp, 7= 13bp, 8= 6bp, 9= 4bp, 10= 4bp, 11= 34bp, 12= 7bp, 13= 19bp, 14= 13bp, 15= 7bp, 16= 3bp. Fig. S10 Sprouting of potato cv Desiree and StCCD8-RNAi excised tuber buds following treatment with water (a) or application of GR24, 2.5 µM (b). The sprouting behaviour of tuber discs from control and RNAi potato lines was monitored over 7 d. Tissue was sampled from mature tubers stored at 4ºC for 10 d after harvest. Values are means of three independent experiments, where each n=20. Error bars represent ±SE (standard error). Statistical differences from the wild type control were calculated with Student’s one-tailed t test, assuming unequal variance. An asterisk was added where there was a significant difference between the calculated (P <0.05) values for RNAi lines and controls. Table S1 The sequence of primers used for cloning of StCCD8 from root tissue StCCD8 5′_597bF: TCACCATACTCTCAAATTCTCTCAA StCCD8 5′_597bpR: GAGAGTAATTTGGCCATGTCACC StCCD8 3′_500bpF: GGAGAATTAGAAGCAGCATTGG StCCD8 3′_500bpR: GCTATTTCAAAAACTGGAAAATTTAG