1465-9921-13-64-S1
advertisement

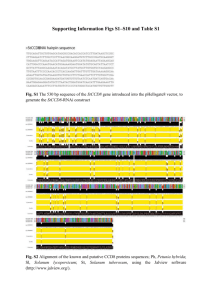
Additional Table 1 List of variants in four FPF genes found in affected family members and sporadic patients. Gene SFTPC SFTPA2 Exon/ Intron intron 1 intron 2 exon 4 intron 4 intron 4 exon 5 exon 6 5'UTR 5'UTR exon 1 exon 2 intron 2 SIFT Polyphen rs4715 rs2070687 rs1124 rs7592 rs17883188 rs17881948 rs2271788 - Predicted protein change NA NA p.Thr138Asn NA NA p.Ser186Asn NA NA NA NA NA NA NA NA tolerated NA NA tolerated NA NA NA NA NA NA - NA NA c.26C>A rs1059046 p.Thr9Asn tolerated exon 3 exon 3 exon 4 exon 4 exon 4 exon 4 exon 4 exon 4 exon 4 c.56C>T c.148G>C c.186G>A c.197C>T c.198A>G c.213G>A c.217A>G c.241G>A c.253C>T rs61862677 rs11554795 rs2434114 rs1713397 - p.Ala19Val p.Val50Leu p.= p.Thr66Ile p.= p.= p.Asn73Asp p.Val81Ile p.Arg85Cys tolerated tolerated NA tolerated NA NA tolerated tolerated tolerated exon 4 c.271G>C rs17886395 p.Ala91Pro tolerated exon 4 intron 4 intron 4 intron 4 intron 4 intron 4 c.282A>G c.292+11C>T c.292+45G>A c.292+46A>C c.292+48dupC c.292+64A>G rs17886221 rs17884130 rs17882071 rs17880662 p.= NA NA NA NA NA NA NA NA NA NA NA Variant rs# c.42+72C>A c.43-21T>C c.413C>A c.435+14G>A c.436-8C>G c.557G>A c.*123G>A c.-120T>G c.-103T>A c.-73G>C c.-35T>G c.-24+62T>C c.[-23-5G>A];[-235G>T] exon 3 intron 2 NA NA benign NA NA benign NA NA NA NA NA NA Allele frequency 0.13 (10/76) 0.33 (19/58) 0.052 (3/58) 0.069 (4/58) 0.12 (7/58) 0.10 (6/58) 0.24 (18/76) 0.93 (106/114) 0.08 (9/114) 0.14 (22/160) 0.91 (42/46) 0.93 (106/114) European allele frequency NA NA 0.23 NA 0.24 0.29 0.30 0.98 0.06 0.86 NA NA NA 0.96 (135/140) NA 0.73 (104/142) 0.82 0.44 (62/142) 0.19 (6/32) 0.78 (86/110) 0.89 (98/110) 0.89 (98/110) 0.89 (98/110) 0.89 (98/110) 0.85 (94/110) 0.86 (95/110) NA 0.95 NA NA 1.00 NA NA NA NA 0.88 (97/110) 0.10 0.86 (95/110) 0.88 (97/110) 0.14 (15/110) 0.14 (15/110) 0.79 (87/110) 0.73 (80/110) 0.02 NA 0.00 0.00 NA 0.00 possibly damaging benign benign NA benign NA NA benign benign benign probably damaging NA NA NA NA NA NA TERT † * TERC intron 4 exon 5 intron 5 intron 5 intron 5 exon 6 exon 6 3'UTR 3'UTR 3'UTR exon 1 exon 2 c.293-68G>A c.342C>T c.370+67C>G c.370+77T>C c.370+77_78dupTA c.420C>T c.667C>A c.*28G>A c.*29A>G c.*120A>G c.-42dupC c.1321_1323delGAG rs1610802 rs61862719 rs1965708 rs17879546 rs17880428 - NA p.= NA NA NA p.= p.Gln223Lys NA NA NA NA p.Glu441del NA NA NA NA NA NA tolerated NA NA NA NA NA exon 4 exon 9 c.1892G>A c.2517G>A - p.Arg631Gln p.= damaging NA exon 10 c.2594G>A rs121918666 p.Arg865His damaging exon 10 c.2648T>G - p.Phe883Cys damaging exon 14 exon 15 exon 16 3'UTR 5'UTR 5'UTR 3'UTR 3'UTR c.3039C>T c.3184G>A c.3324G>A c.*99C>T n.-23G>A n.-11G>A n.*63A>G n.*111C>G rs33954691 rs35719940 rs35033501 rs2853690 rs2293607 - p.= p.Ala1062Thr p.= NA NA NA NA NA NA tolerated NA NA NA NA NA NA NA NA NA NA NA NA benign NA NA NA NA NA possibly damaging NA possibly damaging probably damaging NA benign NA NA NA NA NA NA 0.01 (1/112) 0.07 (8/112) 0.08 (9/112) 0.02 (2/112) 0.10 (11/112) 0.14 (15/112) 0.07 (8/112) 0.02 (2/112) 0.01 (1/112) 0.14 (16/112) 0.01 (1/112) 0.01 (1/112) 0.01 (1/112) 0.04 (2/46) NA NA NA NA NA 0.25 0.11 0.02 0.02 NA NA NA NA NA 0.01 (1/112) NA 0.01 (1/92) NA 0.11 (5/46) 0.01 (1/112) 0.04 (2/46) 0.13 (6/46) 0.02 (1/46) 0.01 (1/108) 0.19 (21/108) 0.02 (1/46) 0.14 0.02 0.04 0.16 NA NA 0.30 NA NA=Not Available; SIFT=Sorting Intolerant From Tolerant; p.= indicates there is no amino acid change for this coding variant † This is a rare known variant previously identified in a normal control and shown to be associated with normal telomere length (Yamaguchi H, Calado RT, Ly H, et al. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N Engl J Med. 2005;352(14):1413-24). * This variant was identified as potentially affecting telomerase function in one study (Calado RT, Regal JA, Hills M, et al. Constitutional hypomorphic telomerase mutations in patients with acute myeloid leukemia. Proc Natl Acad Sci U S A. 2009;106(4):1187-92) and was also identified in control populations; however, it appears to be more frequent in PF and AML populations than controls. We describe this as an unclassified variant as it is in a sporadic PF patient in our cohort. We also do not know the NL population frequency for the variant nor did we perform telomerase length assays.