periodic1-periodicreportsummary
advertisement

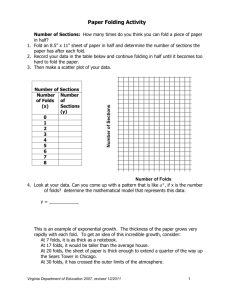
Computational Protein Design has been a breakthrough in redesigning natural proteins to bind new small-molecules and catalyze a variety of non-naturally occurring reactions. The success of this methodology, however, is limited by the stability and evolutionary history of natural proteins. The mutations necessary to place the active site for the function of interest might not be well tolerated by the natural protein and can ultimately lead to problems of protein expression, stability or merely lack of function. To minimize these problems, the methods for designing ideal protein structures1 from scratch are promising. These methods recently implemented in Rosetta and developed by the Baker laboratory have now made possible the de novo design of small and hyperthermostable proteins with different folds at atomic-level accuracy. This major breakthrough now enables control over the protein backbone and should ultimately open the door to the customization of protein structures to the active site of interest. A key of success in this new framework lies in the evaluation of the folding energy landscape of designed proteins by running abinitio folding simulations at rosetta@home (Figure 1A). To date these methods for de novo protein design have been successful in designing protein folds with pockets that are too small to bind a ligand and introduce catalytic activity. Therefore, the next logical step in this field is to expand the current de novo protein design methodology to overcome the limitation of current computational enzyme design methods. The aim of this project is to extend the current de novo computational protein design methodology towards the design of protein folds with cavities of variable size and incorporate binding and/or catalytic functions. Protein folds with curved β-sheets are suitable for this purpose, as they can easily build a deep and accessible cavity, like in jelly rolls, β-barrels or the Nuclear Transport Factor 2 (NTF2) family fold (Figure 1D). By varying the orientation and length of loops and secondary structure elements as well as tuning sheet curvature it is possible to design pockets with variable size and shape. These are folds of higher complexity than those built de novo so far and, indeed, it is a great challenge in itself for the field of de novo protein design. I started this work by extracting different β-sheet design rules from the PDB and modifying the computational design protocol to design protein backbones with curved beta-sheets. As a proof of concept, I computationally designed and experimentally tested the cystatin fold from scratch, which is the minimalist version of a protein with a curved sheet. It consists of an antiparallel β-sheet and one helix. These 72-aminoacid designed proteins were monomeric, thermostable and cooperatively folded (Figure 1B). The structure of two designed proteins was solved by NMR (Figure 1C) and x-ray crystallography, in collaboration with the Northeast Structural Genomics Consortium, and the β-sheet geometry was successfully recapitulated (with 1 Å accuracy with respect to the designed model in both cases). These results validated the approach for the β-sheet design, but the cystatin fold is not suitable for designing active sites yet. I continued working with the more complex the NTF2 fold for this purpose. Four topological variants of this fold have been designed so far with different degrees of complexity (Figures 1E through H). They differ in size (100-122 aminoacids), number of secondary structure elements, types of loop connections and pocket size. In general, these proteins have good expression, are thermostable and mostly monomeric. However, we have only found cooperatively folded designs in only two variants and they are currently under structure determination. B C E F H I Energy (REU) A Rmsd (Å) D G Figure 1. Computationally de novo designed proteins. (A) Example of funnel-shaped energy landscape. (B) Computational model of designed cystatin fold. (C) NMR structure of designed cystatin. (D) Natural NTF2 fold protein. (E-H) Design models of different topological variants of NTF2 fold. (I) Design model of most active variant with a nucleophilic lysine. As a first application of these de novo NTF2 folds, I have designed active sites with a nucleophilic lysine. The activation of protein lysines is required to catalyze aldol and retro-aldol reactions, what are frequently used reactions for the synthesis of new chemicals. In addition active lysines are also interesting to label proteins being monitored in live cells with fluorescence microscopy. For this, the lysine requires to be surrounded by a hydrophobic environment to decrease its pKa and form an enamine with a ketone compound. Depending on the type of ketone compound, the formation of enamine leads to catalysis or covalent binding of the substrate. I computationally designed and experimentally tested active lysine designs in one of the protein variants that were previously characterized. With mass spectrometry four designed proteins with labeling activity were identified, and the most active variant (Figure 1I) has been evolved for labeling function with yeast surface display to higher levels. Interestingly, the computational designs did not have retro-aldol activity, but this evolved variant do shows low levels of enzymatic activity. To our knowledge, this would be the first totally artificial enzyme. The next steps are: (1) finishing the structural characterization of some variants of the NTF2 fold designs, (2) further optimize the designs for retro-aldol activity, (3) design binding for new targets, such as steroid hormones. This work aims at extending the current de novo design capabilities for applications in organic synthesis, image monitoring of proteins in live cells and engineering binders for diagnostics and bioscavengers. 1. Koga N. et al., Nature, 2012, 491, 222