enzyme lab - IB-Biology
advertisement

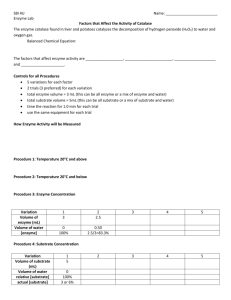
Practical 3: Factors that Affect Enzyme Activity Based on a procedure by J. Smith, Ball State University Learning Targets Explain enzyme catalysis in terms of collision theory, substrate, and active site. Describe the effects of temperature, pH, and substrate concentration on the rate of enzyme activity. Design and carry out an experiment, collect and process data, and report your results. Overview In this experiment, you will use the Vernier Colorimeter with LabPro or LabQuest to measure and calculate the rate of an enzymecatalyzed reaction, and to investigate the effects of pH, temperature, substrate structure, organism of origin, or other variables on reaction rate. Background The enzyme catechol oxidase is found in many plant organs which store food. Examples include potatoes, bananas, and apples. When the organ is cut or damaged by bruising the enzyme’s substrate, catechol, is released from the vacuole. The enzyme catalyzes the partial oxidation of catechol, producing a brownish material called benzoquinone. Quinones are bacterial and fungal inhibitors which may protect the surrounding tissues from infection. In this laboratory exercise, catechol oxidase will be isolated from potatoes. The substrate, catechol, will be mixed with the enzyme. The rate of the enzymatic reaction will be followed by measuring the amount of brown-colored quinone product that is produced over time from the colorless solution of catechol. This treatment will serve as a control, and factors that affect the enzyme such as isomers, inhibitors, pH, and substrate concentration will be studied. Methods and Materials A class set of the enzyme extract will be prepared by Mr. LB. The extract is made by peeling a potato and slicing it into pieces that are put into a blender. A small quantity of distilled water (50-100 ml) is added and the mixture liquefied. The resulting pulp is filtered through cheesecloth. The extract will also be filtered a second time through filter paper to remove starch grains. Solutions of 0.01 M catechol (substrate), hydroquinone (isomer) and resorcinol (isomer) have been prepared. The three chemicals are structural isomers which differ in the position of the hydroxyl groups on the carbon ring as can be seen in the diagrams below: The Vernier Colorimeter is designed to determine the concentration of a solution by analyzing its color intensity. The colorimeter measures the amount of light transmitted through a sample at a user-selectable wavelength. Light from a LED light source passes through a cuvette containing a solution sample. Some of the incoming light is absorbed by the solution and as a result, light of a lower intensity (% Transmittance) strikes a photodiode. The signal is sent to a computer which will display the strength of the signal in the form of a graph of percent Transmittance (%T) or absorbance. This experiment will use %T for recording data. The starting solutions (with substrate) will be clear allowing nearly 100% of the light to be transmitted through the sample tube. If the enzyme produces the colored quinone product the sample will darken, permitting less light to be transmitted through the cuvette. The change in the transmitted light will be directly proportional to the amount of product or quinone formed by the enzymatic reaction. If the %T readings are recorded over time, the rate of the enzymatic reaction can be calculated, because the solution will darken progressively as the reaction proceeds. A “blank” tube will be used calibrate the Colorimeter in order to account for any color in the enzyme extract itself. Therefore, any changes in %T observed by the Colorimeter will be due to the conversion of substrate to product, and not to the color of the enzyme extract. The blank is made by adding 3 ml of distilled water and 4 drops of the enzyme solution to a cuvette. The cuvette should be capped before placing it in the colorimeter. Operation of the Colorimeter and Interfaced Computer 1. Press the < or > button on the Colorimeter to select the 430 nm wavelength setting for use in this experiment. 2. To calibrate, a special cuvette called a “blank” needs to be created (see above). Start the calibration by opening the colorimeter lid. Insert the “blank” cuvette (filled with 3 ml of distilled water and five drops of the enzyme solution). The cuvettes should always be capped when placed in the Colorimeter to prevent liquids from reaching the electronics. Important: Line up one of the clear sides of the cuvette with the arrow at the top of the cuvette slot. Close the Colorimeter lid. 3. Next, press the CAL button to begin the calibration process. Release the CAL button when the red LED begins to flash. The absorbance should now be 0.000 or 0.001. When the LED stops flashing, the calibration is complete. 4. Double click the Y axis label (“absorbance”). You will be asked to choose absorbance, %T, or both as the Y axis. Choose %T. Note - Handle all the cuvettes by the grooved sides to keep fingerprints off the clear sides which can interfere with the transmittance of light. Clean the cuvettes with chem-wipes to remove smudges if necessary. Preparing the Computer for Data Collection Each run in this experiment should last 5 minutes. The timing feature should be set in the experimental set-up so that a reading is recorded every 2 seconds. The Y axis should be in %T and the scale set from 0 to 100. If the data should run off the graph during a test run, remember that the data has not been lost and the "auto scale" icon can be used to rescale the graph. Reaction Mixtures The total volume in the cuvette in all tests is 3 ml. The amount of substrate, isomer and/or distilled water will be altered so that the total is the same in each treatment. The computer must be calibrated and set up for data collection before starting any treatments. All the ingredients except the enzyme extract can be placed into the cuvette. When the computer and the reaction mixture are ready, add the enzyme extract and close the cuvette with a cap. Mix the contents quickly by inverting the capped cuvette then place the it in the Colorimeter as fast as possible and close the lid. Once the cuvette is in place, data collection may begin by clicking on the “collect” button in the upper right corner. Comment - The reaction will begin immediately with the addition of the enzyme. The computer must be preset to start recording the data before the enzyme extract is added to the solution. Cap and invert the cuvette, but do not “shake” the contents because air bubbles will block the light beam and lower transmittance. Investigation Design and conduct an experiment to investigate the effects of pH, temperature, substrate structure, organism of origin, or another variable of your choice on reaction rate. Predict what the results will be before starting a treatment. Connect your prediction to a hypothesis about protein structure and reaction rate (think about what you’ve learned about proteins). You can save the data from each run and add the data from the next treatment to the same screen. This time when you hit the collect button, the program will ask you if it should save the current data. Once you hit OK, the new data collection run will start. You can go as far as the “save current data” step before adding the enzyme to each treatment. Students should note the shape of the graphs produced from the experiment. Is there evidence of a chemical reaction? How does the graph of the treatment compare to the Control? Does the data support your hypothesis on how the treatment affects enzyme activity? Remember to avoid “Three Bears” experimental designs (too much, too little, just right). Use a gradient! Here are some sample research questions about enzyme reactions that you might use as starting points for your investigation. Remember, in each case, you will compare your results to the control treatment as described on the previous page. Can a structural isomer of catechol be used as a substrate? Do structural isomers act as competitive inhibitors? As noncompetitive inhibitors? How do they affect the reaction rate? Are the results the same or different depending on which structural isomer is used? Can you show a significant difference? How does pH affect the reaction rate? What is the optimum pH for this enzyme? How does temperature affect the reaction rate? What is the optimum temperature for this enzyme? How does substrate concentration affect the reaction rate? How does enzyme concentration affect the reaction rate?