tpj12493-sup-0009-Legends
advertisement

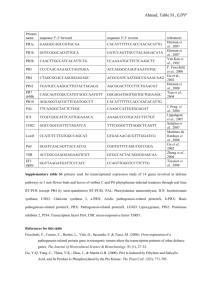
SUPPORTING INFORMATION Figure S1. MicroRNA156 and SPL/SBP genes are conserved in tomato. (a) Expression profiles of miR156-targeted SlySBPs. Heatmap of RNA-seq expression data from flower buds, opened flowers, 1cm and 2cm fruits. The expression values are measured as reads per kilobase of exon model per million mapped reads (RPKM). (b) Sequence alignment of mature miR156 and miR157 in tomato (Sly) and Arabidopsis (At). The EST (LEFL1098BH06) used to generate transgenic tomato plants (Zhang et al. 2011), the mature miR156 sequence (SlymiR156a-h) described in Salinas et al. (2011), and the AtmiR156b mature sequence are shown in bold. nt, nucleotides. (c) stem-loop pulsed RT-PCR analysis to detect AtMIR156b precursor, mature miR156, and SlySBP transcripts in leaf tissues of tomato MT and 156-OE plants. Tomato Tubulin gene (SlTUB) was used as an internal control. Reaction without RT (RT-) and reaction without cDNA (PCR-) are shown as negative controls. Figure S2. miR156 and SlySBP genes regulate tomato vegetative development. (a) Vegetative architecture of adult tomato cv. MT and distinct 156-OE transgenic lines. (b) First (1st) and second (2nd) pair of leaves from adult plants of tomato cv. MT and 156-OE line #18. Figure S3. miR156 overexpression affects tomato fruit development. (a) Representative ripped fruits from tomato MT and distinct 156-OE lines. (b) Detection of miR156 transcripts through stem-loop quantitative pulsed RT-qPCR in ovary and immature fruit tissues of tomato MT and 156-OE lines described in Figure 3. RT-qPCR experiments used fruit and ovary tissues from tomato MT as the reference sample (set to 1.0). Error bars indicate standard deviation of three biological samples. Relative expression levels (R.E.) of miR156 are shown on the top of the graphic bars. Black, gray, and white graphs represent expression data from preanthesis ovaries, postanthesis ovaries, and immature fruits, respectively. Figure S4. Vegetative and fruit phenotypes of weak lines 156-OE#19 and #18 in the Brazilian nondwarf 'Santa Clara' (SC) background. MiR156 overexpression lines were crossed with 'Santa Clara' wild type, and the offspring (harboring the MT mutations in the heterozygous form) were phenotyped. Figure S5. SlySBP genes are down-regulated at variable levels in 156-OE immature fruit tissues. Comparative expression analysis by RT-qPCR of representative SlySBP genes in immature fruit tissues from tomato cv. MT and 156-OE lines. RT-qPCR experiments used tissues from tomato MT as the reference sample (set to 1.0). Error bars indicate standard deviation of three biological samples. R.E., relative expression levels. Figure S6. Computational analysis of putative binding sites for SPL/SBP proteins in tomato genomic sequences. Tomato genomic sequences were retrieved from NCBI and Sol Genomics (http://solgenomics.net/) databases. The sequence GTAC is a core motif essential for efficient SPL/SBP in vitro DNA binding (Birkenbihl et al. 2005). It was therefore used as a driver to identify putative binding sites in specific tomato loci. MC, MACROCALYX; FA, FALSIFLORA. Confirmed binding sites were demonstrated by Yamaguchi et al. (2009). Figure S7. Expression of the MADS-box genes TAG1 and TM29 in developing ovaries of MT and AtMIR156b-overexpressing plants. Black graphic represents gene expression in preanthesis ovaries, while gray graphics represent gene expression in postanthesis ovaries. RT-qPCR experiments used ovary tissues from MT as the reference sample (set to 1.0). Error bars indicate standard deviation of three biological samples. R.E., relative expression levels. Table S1. Oligonucleotide sequences used in this work.