Section 8-1 Answers, p 375

Section 8.1 Questions, page 375
1.
2. You would expect HindIII to result in more fragments because it has a shorter recognition sequence. The longer the recognition sequence of a restriction enzyme, the more specific it will be.
For example, if a restriction enzyme only had a two-base recognition sequence (they typically have 4 to 8), the probability of finding it is one in
4 × 4, or 6 %. If it has an 8-base recognition sequence, it will only locate that sequence one in
4
8 times, or 0.001 % of the time. HindIII has a 6 bp recognition site whereas SmaI has an 8 bp recognition site.
3. Blunt ends are cut straight across the strands, while sticky ends are cut in a zigzag with overhangs.
As the name suggests, sticky ends are generally easier to re-bond because the overhanging base pairs are attracted to one another and form hydrogen bonds.
4. Plasmids can benefit cells by helping them resist antibiotics and toxins such as herbicides and heavy metals.
5. If the cells have been successfully transformed by introduction of the plasmid, they will be resistant to certain antibiotics. Antibiotic resistance indicates that transformation actually occurred.
6. 120 000 ÷ (4 × 4 × 4 × 4 × 4) = 117 cuts
7. Student answers will vary depending on the restriction map they find.
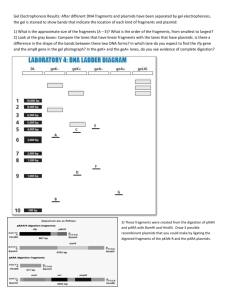
8. (a) (i) 2 fragments; 1236 and 6660 bp
(ii) 2 fragments; 1399 and 6497 bp
(iii) 3 fragments; 429, 1071, and 6396 bp
(iv) 5 fragments; 372, 429, 1071, 1236, and 4788 bp
(v) 7 fragments; 372, 429, 1071, 1056, 1399, 2333, and 1236 bp
(b) BamHI would inactivate the antibiotic resistance because it will interrupt the



![[#SWF-809] Add support for on bind and on validate](http://s3.studylib.net/store/data/007337359_1-f9f0d6750e6a494ec2c19e8544db36bc-300x300.png)