4. Restriction Digest - Size Calculatons
advertisement

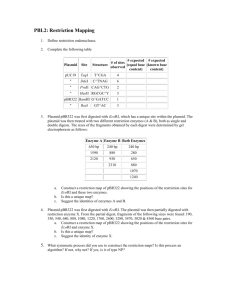
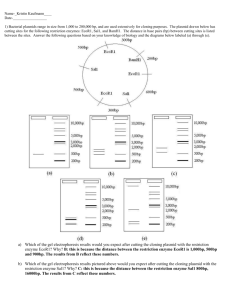
Recognition sequences Restriction enzyme EcoRI cuts the DNA into fragments. Sticky end • Originally protected bacteria from viruses. •Restriction Site = Recognition sequence NOT Cleavage site •Recognition site is a planidrome (just like “radar” “Madam, I’m adam” Example of Restriction Sites: Palindromes: Journal 12.09.10: 1. Which sizes of DNA fragments do you expect to get from the digestion? 2. How can one use the electrophoresis procedure to estimate the size of the DNA fragments? EcoRI HinDIII BamH1 2300 4000 3000 5000 1. Which fragment sizes will result from cutting with: A. HinDIII alone? B. EcoE1 alone? C. Both HinDIII+EcoR1 2. Draw the sizes into the gel. bp 2300 E EcoRI HinDIII BamH1 H B 4000 3000 5000 bp 23000 bp 9000 bp 6000 bp 4000 bp 2300bp 2000 bp 500 bp To Students that are presenting about restriction maps and expected sizes: You may want to peak through the slides about gel electrophoresis, to see the connection between your part and the next – electrophoresis. Restriction digest of pARA-R Recombinant plasmid of interest pARA-R 4720 bp rfp 702bp pARA-R construct Recombinant plasmid of interest pARA-R pARA-R 4720 bp 4720 bp rfp rfp 702bp 702bp Expected fragments pARA-R construct Recombinant plasmid of interest 4720-702=4018bp pARA-R pARA-R 4720 bp 702 bp 4720 bp rfp rfp 702bp 702bp restriction analysis of pARA-R Restriction fragments after digest with Hind III and BamH I BamH I Hind III 4018 bp BamH I Hind III 702bp Additional Resources: Restriction Map Exercise for high-school (if you want to use this)