protocol fragments
advertisement

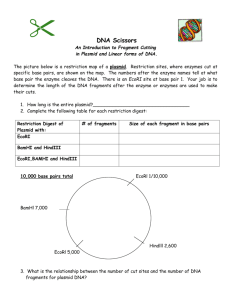
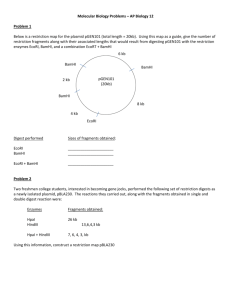
Simulating the effects of three restriction enzymes on Lambda DNA You are presented with a computer programme showing the entire Lambda genome. This enables you to simulate theoretically the same restrictions which you are carrying out practically with the Lambda protocol. 1. Place the disk in the disk drive 2. Open from the disk the file called uncut.doc 3. Browse through the genome 4. Estimate the number of bases in the sequence. (There are two other files on the disk. master.doc is the entire Lambda DNA sequence and serves as a back-up copy of the genome. It is best not to load this so that there is no danger of altering it. The third file simulate - is a copy of this worksheet which can be modified to suit your own teaching needs.) The Lambda base sequence may be cut at various points with restriction enzymes. As we have seen each enzyme cuts within a particular base sequence; e.g. EcoRI cuts between the A and the G in the six base sequence GAATTC and nowhere else. This usually shown as G/AATTC. Other restriction sites are BamHI - G/GATCC and HindIII - A/AGCTT. You can use this programme of the Lambda base sequence to find the restriction sites of these three enzymes - and indeed any other enzymes for which you know the restriction sites. You can then 'cut the genome' and count the number of fragments and the lengths of each of these fragments. These can later be compared with the results of your gel electrophoresis runs from the Lambda protocol. 5. Place the cursor at the beginning of the base sequence 6. Open the EDIT window and select REPLACE 7. In answer to the question 'Find what ?' type in gaattc 8. Move the cursor to 'Replace with ?' and type in G^pAATTC 9. Select 'Replace all' This cuts the base sequence at all the EcoRI restriction sites and puts a paragraph break (^p) at each point where the sequence has been cut. It also converts the recognition sequence of six bases to upper case letters so that you can easily see them. Questions 1. How many restriction sites were there ? 2. How many resulting fragments will there be ? 10. Choose 'OK' and then choose 'close' 11. Highlight the first 'paragraph' of the genome (in reality the first fragment resulting from restriction) by double clicking anywhere in the margin alongside the first few lines 12. Open the TOOLS window and select WORDCOUNT. Note the number of characters (i.e. the number of bases) in this fragment and enter this number alongside the i) below the EcoRI bar below: EcoRI i)----------------------------------------ii)------------iii)----------iv)------------------v)---------------vi)-----------13. Now select 'cancel' 14. Move the cursor to the start of the next paragraph, highlight as before, and repeat - noting the number of characters (i.e. bases) alongside the ii) below the bar 15. Repeat this process until you reach the end of the genome You should now have the number of fragments you noted in answer to the second question above and shown on the EcoRI bar. You should also have the fragment lengths in terms of the number of bases. 16. Now arrange the fragments resulting from the restriction by EcoRI in order of size, starting with the largest. Enter these in the table below: 17. Close the file remembering to answer 'No' to the question 'Do you want to save changes to uncut.doc ?' 18. Repeat the whole process for BamHI and HindIII, in each case entering the fragment sizes below under the appropriate bar BamHI - restriction site g/gatcc i)----------------ii)-----------------------------iii)------------iv)--------------v)-------------------vi)----------------- Hind III - restriction site a/agctt i)---------------------------------------ii)-----iii)-----iv)-----------------v)-----vi)-----vii)------------viii)---------- 19. Close the file remembering to answer 'No' to the question 'Do you want to save changes to uncut.doc ?' 20. Now arrange your fragments resulting from the restriction by BamHI and HindIII in order of size, starting with the largest and enter these in the table below alongside the values for EcoRI: EcoRI largest fragment BamHI HindIII smallest fragment total number of fragments 21. Compare all three sets of fragments in the table with your gels and identify as many of the bands as you can.