Enzyme Kinetics
advertisement
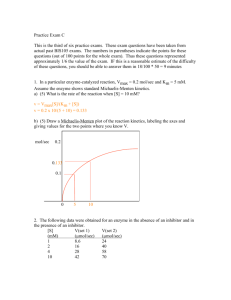
ENZYMES KINETICS, INHIBITION, REGULATION Muhammad Jawad Hassan Assistant Professor Biochemistry Michaelis-Menten kinetics V0 varies with [S] Vmax approached asymptotically V0 is moles of product formed per sec. when [P] is low (close to zero time) E + SESE + P Michaelis-Menten Model V0 = Vmax x[S]/([S] + Km) Michaelis-Menten Equation Steady-state & pre-steady-state conditions At equilibrium, no net change of [S] & [P] or of [ES] & [E] At pre-steady-state, [P] is low (close to zero time), hence, V0 for initial reaction velocity At pre-steady state, we can ignore the back reactions Michaelis-Menten kinetics (summary) Enzyme kinetics (Michaelis-Menten Graph) : At fixed concentration of enzyme, V0 is almost linearly proportional to [S] when [S] is small, but is nearly independent of [S] when [S] is large k1 k2 Proposed Model: E + S ES E + P ES complex is a necessary intermediate Objective: find an expression that relates rate of catalysis to the concentrations of S & E, and the rates of individual steps Michaelis-Menten kinetics (summary) Start with: V0 = k2[ES], and derive, V0 = Vmax x[S]/([S] + Km) At low [S] ([S] < Km), V0 = (Vmax/Km)[S] At high [S] ([S] > Km), V0 = Vmax When [S] = Km, V0 = Vmax/2. Thus, Km = substrate concentration at which the reaction rate (V0) is half max. Range of Km values Km provides approximation of [S] in vivo for many enzymes Lineweaver-Burk plot (double-reciprocal) Allosteric enzyme kinetics Sigmoidal dependence of V0 on [S], not Michaelis-Menten Enzymes have multiple subunits and multiple active sites Substrate binding may be cooperative Enzyme inhibition A competitive inhibitor Methotrexate A competitive inhibitor of dihydrofolate reductase - role in purine & pyrimidine biosynthesis Used to treat cancer Kinetics of competitive inhibitor Ki = dissociation constant for inhibitor Increase [S] to overcome inhibition Vmax attainable, Km is increased Competitive inhibitor Vmax unaltered, Km increased Kinetics of non-competitive inhibitor Increasing [S] cannot overcome inhibition Less E available, Vmax is lower, Km remains the same for available E Noncompetitive inhibitor Km unaltered, Vmax decreased Enzyme inhibition by DIPF Group - specific reagents react with R groups of amino acids diisopropylphosphofluoridate DIPF (nerve gas) reacts with Ser in acetylcholinesterase Affinity inhibitor: covalent modification Catalytic strategies commonly employed 1. Covalent catalysis. The active site contains a reactive group, usually a nucleophile that becomes temporarily covalently modified in the course of catalysis 2. General acid-base catalysis. A chemical reaction is catalyzed by an acid or a base. The acid is often the proton and the base is often a hydroxyl ion. A molecule other than H2O may play the role of a proton donor or acceptor. 3. Metal ion catalysis. Metal ion can function in several ways; • can serve as an electrophile, stabilizing a negative charge on a reaction intermediate. • can generate a nucleophile by increasing the acidity of a nearby molecule, such as H2O in the hydration of CO2 by carbonic anhydrase. • can bind to substrate, increasing the number of interactions with the enzyme. 4. Catalysis by approximation. Bringing two substrates together along a single binding surface on an enzyme Enzyme specificity: chymotrypsin Cleaves proteins on carboxyl side of aromatic, or large hydrophobic amino acid Bonds cleaved, indicated in red The enzyme needs to generate a powerful nucleophile to cleave the bond A highly reactive serine (#195) in chymotrypsin 27 other serines not reactive to DIPF, Ser 195 is a powerful nucleophile DIPF: di-isopropylphosphofluoridate, only reacts with Ser 195 Covalent catalysis Hydrolysis in two stages Acylation to form acyl-enzyme intermediate Ser 195 OH group attacks the carbonyl group Deacylation to regenerate free enzyme Acyl-enzyme intermediate is hydrolysed Chymotrypsin in 3D 3 chains; orange, blue, & green Catalytic triad of residues, including Ser 195 2 interstrand, & 2 intrastrand disulfide bonds See Structural Insights Synthesized as chymotrypsinogen Proteolytic cleavage to 3 chains The catalytic triad (constellation of residues) Ser 195 converted into a potent nucleophile, an alkoxide ion Asp 102 orients His 57 Imidazole N as base catalyst, accepts H ion, positions & polarizes Ser H ion withdrawal from Ser 195 generates alkoxide ion Regulatory Strategies: Enzymes & Hemoglobin 1. Allosteric control. Proteins contain distinct regulatory sites and multiple functional sites. Binding of regulatory molecules triggers conformational changes that affect the active sites. Display cooperativity: small [S] changes - major activity changes. Information transducers: signal changes activity or information shared by sites 2. Multiple forms of enzymes (isozymes). Used at distinct locations or times. Differ slightly in structure, in Km & Vmax values, and in regulatory properties 3. Reversible covalent modification. Activities altered by covalent attachment of modifying group, mostly a phosphoryl group 4. Protleolytic activation. Irreversible conversion of an inactive form (zymogen) to an active enzyme Aspartate transcarbamoylase reaction Committed step in pyrimidine synthesis: inhibited by end product CTP CTP inhibits ATCase CTP stabilizes the T state CTP binds to regulatory subunits R and T states in equilibrium ATCase displays sigmoidal kinetics Substrate binding to one active site converts enzyme to R state increasing their activity: active sites show cooperativity Basis of sigmoidal curve R & T states equivalent to 2 enzymes with different Kms Cooperativity Effect of CTP on ATCase kinetics CTP stabilizes the T state, curve shifts to right Effect of ATP on ATCase kinetics ATP, allosteric activator, stabilizes R state, curve shifts to left Oxygen delivery by hemoglobin, cooperativity enhanced 98 - 32 = 66% 63 - 25 = 38% Cooperativity enhances delivery 1.7 fold Partial pressure of oxygen Heme group structure 4 linked pyrrole rings form a tetrapyrrole ring with a central iron atom. side chains attached Position of iron in deoxyhemoglobin Iron slightly outside porphyrin plane His (imidazole ring) binds 5th coordination site 6th site for O2 binding O2 binding, conformational change Iron moves into plane, his is pulled along Quaternary structure of hemoglobin Pair of identical alpha-beta dimers Transition from T-to-R state in hemoglobin Interface most affected As O2 binds, top pair rotate 15o with respect to bottom pair Oxygen affinity of fetal v maternal red blood cells Fetal Hgl does not bind 2,3-BPG, higher O2 affinity Fetal hemoglobin tetramer has 2 alpha & 2 gama chains, Gene duplication Isozymes of lactate dehydrogenase: glucose metabolism Rat heart LDH isozyme profile changes with development H(heart) isozyme (chain)= square, M(muscle) isozyme = circle Tissue content of LDH Functional LDH is tetrameric, with different combinations of subunits possible. H4 (heart) has higher affinity for substrates than does M4 isozyme, different allosteric inhibition by pyruvate H4 H 3M H2M2 HM3 M4 Some isozymes in blood indicative of tissue damage, used for clinical diagnosis Increase in serum levels of H4 relative to H3M, indicative of myocardial infraction (heart attack) Examples of covalent modification Phosphorylation widely used for regulation Gamma phosphoryl group Some known protein kinases Protein phosphotases Reverse the effects of kinases, catalyze hydrolytic removal of phosphoryl groups attached to proteins Activation by proteolytic cleavage Secretion of zymogens by acinar cell of pancreas Pancreas, one of the most active organs in synthesizing & secreting proteins Acinar cell stimulated by hormonal signal or nerve impulse, granule content released into duct to duodenum Proteolytic activation of chymotrypsinogen Active enzyme generated by cleavage of a single specific peptide bond 3 chains linked by 2 interchain disulfide bonds, (A-B & B-C) Conformations of chymotrypsinogen & chymotrypsin Electrostatic interaction between Asp 194 carboxylate & Ile 16 -amino group possible only in chymotrypsin, essential for activity Zymogen activation by proteolytic cleavage Zymogens orange, active enzymes yellow Secreted by cells that line duodenum Digestive proteins of duodenum Interaction of trypsin with its inhibitor Lys 15 & Asp 189 form salt bridge inside the active site