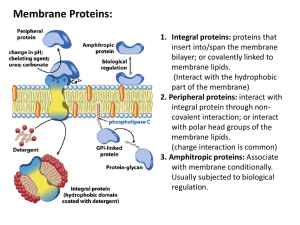
Catabolism vs Anabolism
advertisement

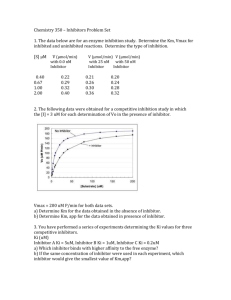
BIBC 102 Metabolic Biochemistry! Lectures #1 and #2!! • Hello. I am Tommy Wootton. • Feel free to shut me up any time for questions! Catabolism vs Anabolism To break down (AKA Cut-abolism!) To build up Anabolic steroids! know the 20 amino acids by name, three letter code, one letter code, and be able to recognize (as opposed to draw) the structures. Amino acids: The Molecular Tool Box (G) (L) (D) (M) (I) (S) (P) (T) (N) (C) (Q) (E) (K) fig3-5 (V) (A) (R) (H) (F) (Y) (W) Amino Acid/Peptide Bond Protein Structure Primary Structure - AA sequence Secondary Structure - 3D structures! ( helix, sheets) Tertiary Structure - Interactions between these secondary structures Quaternary Structure - Multiple subunit interactions So, lets use a word analogy… “This guy SUCKS!” -Hopefully NOT any of you Primary? --> letters Secondary? --> words Tertiary? --> sentence Quaternary? --> multiple sentences interacting to achieve satisfactory expression of distaste Energy map of a reaction fig6-2 DG‡ is the activation energy Activation energy and reaction rate fig 6-3 4 Main Enzymatic Catalytic Mechanisms Entropy Reduction Acid Base Catalysis Metal Ion Catalysis Covalent Intermediates Entropy Reduction Bringing the pieces together! Forcing them to interact. Think of Randy’s analogy with 2 cats. Or a cat and a dog. If they’ve got plenty of space, they’re not going to interact. But if they’re brought together… Acid Base Catalysis Providing or picking up protons to allow for more efficient reaction catalysis. Happens in Chymotrypsin! H+ provided by Ser195 OH group! Metal Ion Catalysis Involves the stabilization of intermediate structures through ionic bonding from metal ion cofactors like Zn2+. Haven’t talked about it much, but some enzymes do it. Covalent Intermediates When a substrate forms a covalent bond with the enzyme to form an intermediate! Remember TPP (Thiamine Pyrophosphate) or the dihydrolipoyl arm from Pyruvate Dehydrogenase Complex?! Sample Problem! free energy reaction Ligand Binding The dissociation constant (Kd) S + E (Separate) SE (bound together!) Kd [S][E] = ____ [SE] SO… If Kd is BIG, S and E tend to be separate (HIGH dissociation, get it?) If Kd is little, S and E will tend to be bound So back to ligand enzyme binding equation… If Kd = 0, LB = B (Saturation! Every emzyme is bound) BUT, if Kd is big (=enzyme and ligand prefer to be separate), LB becomes a smaller proportion of B What if Kd = L? Hmmm… Now something cool… If Kd = L, then LB = 1/2 [B] So it can tell you (and let you compare!) the affinities of an enzyme to a ligand in different conditions… Related to the Michaelis-Menton Equation? Yep! Pretty similar. Except Michaelis-Menton represents enzymatic rates, NOT ligand binding Vo = Vmax ( ) [S] ______ [S] + Km LB = B ( ) [L] _____ [L] + Kd Same general equation! So, same general shape… So really, just a change in axes labeling! - Km = efficiency of RXN - Kd = efficiency of binding - Gives info about enzyme activity -Gives info about ligand binding What about a cooperative enzyme? Due to quaternary structure. Multiple subunits and binding afinity increases as each is bound. Makes Vo increase as [S] increases (until you approach Vmax and saturation. Ex. Heme vs. Myoglobin So how fast can an enzyme work? Vmax = Kcat [E]total Whats Kcat? The rate at which an individual enzyme can undergo catalytic function. Essentially it’s how quick each individual enzyme molecule can do it’s work. Sample Problem! M is half saturated when S equals the value of Km… 5 uM S/(S + Km) = 20/25. So 0.8, or 4/5, or even 80% are acceptable L is half saturated when S equals the value of Km… 20 uM The ratio is 1. Since the kcats are the same, the maximal rates (for a given amount of enzyme) are the same. Three Types of Inhibitors! 1. Competitive Inhibitor 2. Uncompetitive Inhibitor 3.“Suicide” Inhibitor Competitive Inhibitors *not quite like this! The dumb@$$ in this picture would be a dysfunctional ligand… BUT it’s about the concept. They take up space AT the active site. What does this look like molecularly? One more view… Uncompetitive Inhibitors You’re almost there. At the computer (AKA active site). Ready to go, but someone (inhibitor) is bugging ya. An uncompetitive inhibitor binds away from the active site. Doesn’t compete for the active site. Not like in this pic. Looks like? Another view… “Suicide” Inhibitor Literally takes the computer (enzyme) out of commission. Covalently binds to the enzyme and breaks it down so it doesn’t work anymore. -Lowers Vmax. -No effect on Kcat :( Allosteric regulation Allo = other, steric = site SO, regulation through binding at a site OTHER than the active site. Can be inhibitors OR activators. Often the regulator is a product or substrate (allows for negative feedback!) Chymotrypsin A serine protease! Cuts up proteins! Hydrolyzes its substrates (W, Y, F, L) at the carboxy terminal of the peptide. W=Tryptophan, Y=Tyrosine, F=Phenylalanine, L=Leucine… The Catalytic Triad = Asp102, His57, and Ser195. Asp102 stabilizes the positive charge of His57 when it picks up proton from Ser195 after nucleophilic attack. The Catalytic Triad = Asp102, His57, and Ser195. Hydroxy group of Ser 195 attacks. Proton transferred to His57. Asp102 stabilizes the positive charge of His57 when it picks up proton from Ser195 after nucleophilic attack. Lineweaver-Burk Plots! A couple things to remember: -Axes are reciprocals -Tells you Vmax, Km Know where they are! -Be able to think about it. Talk through it. Shouldn’t just look like random lines! Competitive Inhibitors! As [I] increases, Km increases, but Vmax stays the same! This is because you will need more S to reach 50% saturation (to displace I), but the max rate can theoretically still be reached! Just takes more S. Uncompetitive Inhibitors Here, Vmax decreases as [I] increases. Also, Km decreases! This is because I binds to an allosteric site and while bound, makes E inactive. Sample Problem! 1/[S] 1/Vo -1/Km 1/Vmax When Michaelis-Menton enzymes are plotted in this manner, the resulting data is a straight line, making it easier to see when an enzyme behaves this way and easier to discern what kind of inhibitor is involved in an experiment.