Quantitative Analysis of Enzyme Activity (PowerPoint) Northeast 2014
advertisement
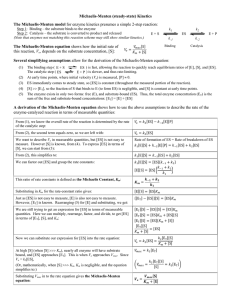
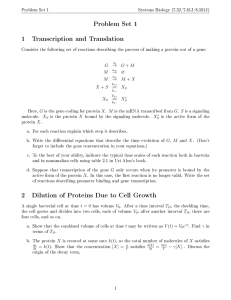
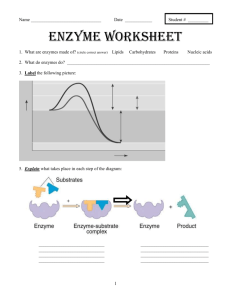
Group 1 – Interface of Chemistry and Biology Quantitative Analysis of Enzyme Activity Leon Dickson Howard University Steven Glynn Stony Brook University Lindsay Hinkle Harvard University Kevin Jones Howard University Scott Sutherland Stony Brook University Rosa Veguilla Harvard University Goals and Objectives Learning Goal: Students will have the ability to manipulate, interpret, and produce visual representations of data describing kinetic properties of enzymes Learning Objectives: Students will be able to: • Determine reaction rates from experimental time-course data • Produce the Michaelis-Menten plot from experimental data • Interpret changes in reaction conditions from different Michaelis-Menten plots • Design an experiment to generate data for a Michaelis- Who you are: Upper level Biochemistry major who has completed Calculus and Introductory Chemistry and Biology We’re halfway through a lecture in steady-state enzyme kinetics. See tip sheet for topics you have covered. HIV-1 protease is crucial for the replication of HIV (necessary for HIV replication) Inhibiting the activity of HIVI protease is a strategy for combating the virus The first step in designing an inhibitor is to understand the kinetic properties of the enzyme Steady-state enzyme kinetics Assumptions of Michaelis-Menten kinetics: 1. The reaction is at equilibrium 2. The reaction is at steady-state Choose the components of the HIV-1 protease reaction HIV-1 protease Viral polypeptide HIV-1 protease/ Viral polypeptide complex Cleaved viral polypeptides Initial reaction velocity (μM sec-1) An enzyme’s response to substrate can be visualized using the Michaelis-Menten plot Vmax Vmax/2 KM Substrate concentration (μM) MichaelisMenten Equation Activity 1 Match the experimental data to the corresponding line on the plot of timecourse reactions Remember that the slope of the time-course corresponds to the rate of the reaction at a given substrate concentration Clicker question Using your handout, identify which time-course corresponds to an initial [S] of 25 uM? Activity 1I A. Use the reaction velocities from the time-course data to construct a Michaelis-Menten plot B. Use your plot to estimate Vmax and KM for your enzyme Vmax Vo KM [S] Clicker question What value for KM did you determine from your MichaelisMenten plot? A. 0– 5 μM B. 8 – 12 μM C. 40 – 50 μM D. 80 – 100 μM Here’s what it should look like: Using enzyme kinetics to evaluate drug candidates -Group1avir Vmax = 96.4 μM KM = 10.2 μM + Group1avir Vmax = 96.4 μM KM = 47.0 μM Is Group1avir a possible drug candidate against HIV? Trends in Annual Age-Adjusted* Rate of Death Due to HIV Infection, United States, 1987−2009 Saquinavir released onto market by Roche Note: For comparison with data for 1999 and later years, data for 1987−1998 were modified to account for ICD-10 rules instead of ICD-9 rules. *Standard: age distribution of 2000 US population In the next lab session you will: • Measure rates of an enzymecatalyzed reaction • Use your data to construct a Michaelis-Menten plot • Determine values for Vmax and KM Let’s remind ourselves what we’ve accomplished In this class you: • Determined a reaction rate from experimental timecourse data • Produced the Michaelis-Menten plot from experimental data and estimate the kinetic parameters • Used Michaelis-Menten plots to infer changes in enzyme activity, e.g. in the context of a human disease