Molecular and Cellular Biophysics course I & II » 2005/2006

Questions for « Molecular and Cellular Biophysics I & II 2005/2006 »
1. Structure of proteins
1.1 Poly-L- leucine in an organic solvent such as dioxane is
-helical whereas poly-
Lisoleucine is not. Why do these amino acids with the same number and links of atoms have different helix-forming tendecies?
Note: Look to Fig 3.28 in (1).
1.2 Why do the amino acids proline and glycine have in proteins helix-breaking properties and often occur in
-turn structures?
1.3 Describe major chemical and structural properties of a peptide bond and the consequences for regular protein structure formation.
Note: Read chapters 3.2.2 & 3.3 in (1).
1.4 Identify the groups in a protein that can form hydrogen bonds with an arginine side chain at pH 7.
Note: Read chapter 3.1 in (1).
1.5 For an amino acid like alanine, the major species in solution at pH 7 is the zwitterionic form. Assume a pK value of 8 for the amino group and a pK value of 3 for the carboxylic group; estimate the concentrations of positively charged amino acid species (carboxylic & amino groups protonated), zwitterionic species, and negatively charged species (carboxylic & amino groups deprotonated).
Note: Use Henderson Hasselbach equation.
1.6 A solution of a protein whose sequence includes 3 tryptophan, 6 tyrosine residues, and no phenylalanine residues has an absorbance of 0.15 at 280 nm in an optical cuvette with a pathlength of 1 cm. Estimate the concentration of the protein in molarity. If the protein has a molecular mass of 100 kd, estimate the protein concentrationin mg protein per mililiter of solution. This is an important practical problem occuring biochemistry laboratories when you work with purified proteins.
Note: Use spectra in Fig 3.11 of (1) for extinction coefficients.
1.7
Describe major energetic contributions for protein folding into regular structures
1.8 (a) Explain the hydrophobic effect.
(b) Explain in this context the so-called hydropathy index, how this index is established, and how it can be used for predicting protein folding into regular structures.
(c) Explain the differences of protein folding between water-soluble and membrane proteins.
(d) What are the major structural determinants for regular structures in the bilayer spanning segments of membrane proteins ?
Note: Chapter on membranes in (1); Chapter: « Structure of Proteins » in (3)
2. Methods for exploring proteins
2.1 What is the largest possible value for the diffusion coefficient that a molecule of
M = 50000 dalton can have at 20 o C (viscosity
= 0.01 poise, partial molar volume
=
0.73 mL/g)
Note: Read introductory part of chapter on sedimentation in (3) & ch 3 of (1).
2.2 Centrifugation experiment: The data in the table below describe the variation of the sedimentation coefficient s, and the diffusion coefficient D for a protein as a function of pH. Expélain what happens to the protein at low and high pH, assuming that it is in its native state between pH 5-7. pH s x 10 13 [sec]
2
3
4
5
2.93
3.02
3.89
4.41
D x 10
7.91
8.00
8.00
5.90
7 [cm 2 /sec]
6
7
8
4.40
4.15
3.60
5.92
5.61
4.86
9
10
2.25
2.20
3.08
2.97
Note: Read introductory part of chapter on sedimentation in (3) & ch 3 of (1).
2.3 (a) A protein with partial molar volume of 0.72 mL/g is studied by sucrose density gradient centrifugation at 5 oC. If the gradient runs from 10 to 30% sucrose, by what percent will the sedimentation coefficient decrease as the protein proceeds from the meniscus to the bottom of the tube? The following data for sucrose solution at 5 oC are required:
% sucrose density [g/mL] viscosity
[centipoise]
10
30
1.0406
1.1315
2.073
4.422
(b) If the meniscus is 8.0 cm, and the bottom of the tube is 16.0 cm from the center of rotation, by what percentage will the velocity of sedimentation change in traversing the tube ?
Note: Read introductory part of chapter on sedimentation in (3) & ch 3 of (1).
2.4 An invertebrate hemoglobin is found, under native conditions, to have a sedimentation coefficient of about s = 4.4 x 10
6x10
-7
[cm
2
/sec] (20 o
-13
[sec] and a diffusion coefficient of D =
C, water). The partial molar volume
is estimated to be 0.73
[cm
3
/g]. The following data are found from SDS-gel electrophoresis :
- After treatment with
-mercaptoethanol, the protein migrates as a doublet. The bands have traveled 10.0 and 10.6 cm.
- In the absence of
-mercaptoethanol, only the 10.0 cm band is seen, but there is also anew band at 5.6 cm.
- A series of standard proteins on the same gel migrate as follows:
Protein M [g/mol] d [cm]
Phosphorylase b
Bovine albumin
94000
67000
0.5
1.1
Ovalbumin 43000
Carbonic anhydrase 30000
Trypsin inhibitor 20100
-lactalbumin 14400
3.9
6.6
9.3
11.7
Describe the subunit structure of this protein. (Don’t expect it to behave like human hemoglobin; invertebrate hemoglobins are often quite different from the mammalian types!)
Note: Read introductory part of chapter on sedimentation in (3) & ch 3 of (1).
2.5
Describe principles of SDS gel electrophoresis and of isoelectric focussing and for which purpose these two methods are used.
MALDI-TOF masspectroscopy is recently used in combination with 2D electrophoresis. Explain the basics of this technique, what kind of information can be obtained and which role it plays in the field of proteomics.
2.6
What are the principal techniques to determine high resolution 3D structures of biopolymers (proteins, nucleic acids)? Describe their principles.
2.7
There are a number of spectroscopic tecghniques which deliver structural information about proteins such as Circular Dichroism spectroscopy and FTIR spectroscopy. Describe the basic principles of these techniques and the type of information one can obtain for determining the structure of proteins.
2.8
Figure 10.15 in ref. (3) show typical CD spectra of polypeptides in a helical, beta strand and random conformation. Pick 3 wavelengths that would discriminate most sensitively among these forms. Describe how you might analyze the data for an unknown protein in terms of CD values at these three different wavelengths.
2.9
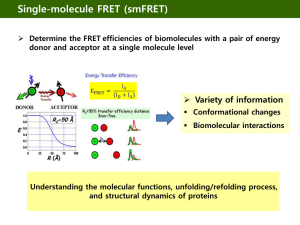
Describe the basic principles of fluorescence resonance energy transfer (FRET) and some typical applications of this technique.
2.10
A protein is labeled by 2 chromophores suited to perform FRET. The typical
Förster distance is R
0
= 2.3 nm. The measured FRET efficiecy is found to be
0.015. Estimate the distance between the 2 labels.
2.11
Describe basic principles of fluorescence anisotropy. What can be learned using this technique?
2.12
Muscle contracts by sliding myosin filaments relative to actin filaments. The myosin headgroup hydrolizes ATP to get the energy for its globular motor domain to move a long lever arm. A recent publication reports on a FRET experiment, where the green fluorescent protein GFP and its mutated blue fluorescent protein
BFP was used to demonstrate that during the working stroke the lever arm tilts against the motor domain. Experimental outline : GFP was attached to the Nterminus of the myosin, and BFP was attached to the C-terminus. Exciting BFP causes the GFP to fluoresce with an efficiency of FRET transfer of 33% after ATP is hydrolized to ADP, which corresponds to a distance of 3.8 nm. Before the hydrolysis, the FRET efficiency is 0.082. How far do the two chromophores move relative to each other during the working stroke?
2.13
Describe 3 different methods of labeling proteins with fluorophores and discuss the advantages/disadvantages of them when compared to each other.
2.14
Surface sensitive techniques such as surface plasmon resoonance (SPR) and total internal reflection fluorescence (TIRF) are suited to determine molecular interactions. Describe the basic principles of these techniques and their principal application.
2.15
The R0 values for the fluorophores used in many biological FRET experiments are about 6 nm. Using this value: How far apart do the donor and acceptor have to be for the FRET efficiency to drop to 10% of its maximum value?
2.16
An RNA molecule that can exist in two different structural states: either in a compact, folded conformation or an open conformation. The two states are probed by singler molecule FRET measurements. For the fluorophore used, R0 = 6.5 nm.
(a) The efficiency of FRET is observed to fluctuate erratically between 0.9 and
0.2. Estimate the distances between the donor – acceptor pair for the two structural states.
(b)
Data on « dwell time » in the high FRET efficiency (folded) state have been accumulated for long observation times on a number of molecules:
Average dwell time
0.1
0.2
0.5
0.6
0.9
1.1
1.2
Number of events
33
23
18
14
9
6
4
Calculate the rate of unfolding from these data. Is it a first-order process?
Hint: Plot number of events as function of dwell time, calculate relaxation time
, the reaction rate constant k = 1/
This exercise is quite instructive what you can learn from single molecule experiments which would be hidden in ensemble measurements
2.17
Novel techniques have been used recently to investigate the behavior of single molecules. Describe a few briefly and explain what are the novel information one can obtain from single molecule techniques one cannot get from ensemble measurements.
2.18
Describe the basic principles of fluorescence correlation spectroscopy and what are the major applications of this technique.
3. Equilibrium binding experiments
3.1 The subunits of hemocyanin from a particular species can undergo the following association reaction.
2P = P
2
2 P
2
= P
4
K
K
12
24
= [P
= [P
2
4
]/[P]
]/[P
2
2
] 2
K
14
= [P
4
]/[P]
4
The equilibrium constants have been measured as afunction of temperature T:
T [ o C] K
12
K
14
4
10
15
30
1.3 x 10
7
1.2 x 10
7
1.5 x 10
7
---------
4.5 x 10
20
1.0 x 10
21
3.0 x 10
21
4.0 x 10
22
All the concentrations are expressed in mole/liter.
Estimate
H 0 ,
S 0 , and
G 0 at 20 o C for the reactions.
3.2
The data below show the binding of oxygen to the sqid hemocyanin. Determine from a Hill plot whether thebinding is cooperative and estimate the number of subunits in the molecule p(O2)
1.13
5.55
7.72
10.72
% saturation
0.3
1.33
1.92
3.51 p(O2)
136.7
166.8
203.2
262.2
% saturation
55.7
67.3
73.4
79.4
31.71
71.87
100.5
123.3
8.37
18.96
32.90
47.80
327.0
452.8
566.9
736.7
83.4
87.5
89.2
91.3
Textbooks:
(1) JM Berg, JL Tymoczko & L Stryer:
Biochemistry
5th edition, Freeman, 2002
(2) CR Cantor & PR Schimmel:
Biophysical Chemistry, vols 1-3
Freeman, 1980
(3) KE van Holde, WC Johnson & PS Ho:
Principles of Physical Biochemistry
Prentice-Hall, 1998