Proteomics investigation into cardiac endothelial
advertisement

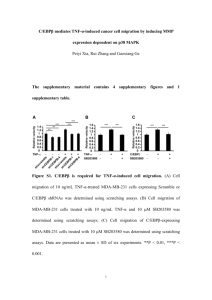
Proteomics investigation into cardiac endothelial cells using the Orbitrap at the Proteomics facility of the University of Stellenbosch Salome Smit Central Analytical Facility University of Stellenbosch Overview 1. Proteomics analysis of cardiac endothelial cells 2. SILAC experiment with HIV-1Tat protein 3. Summary CARDIAC MICROVASCULAR ENDOTHELIAL CELLS = CMECs • Vascular endothelium long thought to be a mere selectively permeable barrier between the circulation and sub-endothelial tissues, is now known to be a master regulator of vascular homeostasis, • Controlling functions such as vasomotor activity, thrombosis, inflammation and redox balance • When endothelial function becomes compromised as observed in cardiovascular risk conditions such as diabetes mellitus, vascular homeostasis is lost resulting in increased oxidative stress, a loss of nitric oxide (NO) bioavailability, increased endothelial cell expression of pro-inflammatory vascular adhesion molecules and increased endothelial permeability. •These pathophysiological changes underlie the phenomena of endothelial activation and endothelial dysfunction, of which the latter in particular is regarded as the early forerunner of atherosclerosis. All slides for CMEC work courtesy of Prof Hans Strijdom, University of Stellenbosch CMECs • In the heart, the myocardial capillaries (leading to ischaemic heart disease) are made up of cardiac microvascular endothelial cells (CMECs). • CMECs show distinct structural and functional adaptations compared to other endothelial cell phenotypes in view of their location in the myocardium where they are closely associated with surrounding cardiomyocytes. • There is intimate CMEC-cardiomyocyte arrangement • cardiomyocytes are regarded as the primary cellular recipients of paracrine messengers secreted by CMECs, such as NO and endothelin-1. CMECs • CMECs: PIVOTAL ROLE IN BOTH MYOCARDIAL FUNCTION AND INJURY • Optimal diffusion of oxygen and nutrients • Reciprocal signalling with cardiomyocytes • Regulate cardiomyocyte growth and development • Regulation of cardiomyocyte contractile function & rhythmicity • Therefore, CMECs are now recognized as important regulators of myocardial function. TNF-α: RELEASE, BINDING AND EFFECTS Inflammation; Tissue Injury (eg Ischaemia); Aging; Cardiovascular Risk Factors (Obesity; DM); Heart Failure ↑TNF-α Release TNF-R1 APOPTOSIS INFLAMMATORY RESPONSE / PRO-SURVIVAL TNF-R2 ANTI-APOPTOSIS / ANTI-NECROSIS Vascular endothelial cells are the PRIMARY targets of circulating TNF-α (Pober 2004); Express both TNF-R1 and TNF-R2 (Madge 2001) What is the effect of TNF-α on CMEC? What would be a novel and optimal method to study these cells? To gain the most information METHODS: LARGE-SCALE PROTEOMICS: SDS-PAGE IN-GEL TRYPSINISATION NANO LIQUID CHROMATOGRAPHY MASS SPECTOMETRY PROTEIN ID • Relatively few papers measure large-scale protein expression and regulation in vascular endothelial cells of any type (“only” 350 since 2001): Surprising! (Richardson 2010) •Pubmed search: <5 papers reported on any form of proteomic analysis performed on CMECs PROTEIN REGULATION: Control Down 77 Control Shared: 1056 TNF-α Up 143 TNF-α Maxquant™: 1102 proteins Control Down 226 TNF-α Up 269 Shared: 1214 Control TNF-α Sieve™: 1511 proteins TNF-α: 5ng / ml ; 24h UP REGULATED AND TNF-α ONLY: 16 proteins DAVID Bioinformatics Resources® UP REGULATED AND TNF-α ONLY: 51 proteins MITOCHONDRIAL PROTEINS: • ATP Synthase subunits (TNF only); • Acetyltransferase component of Pyruvate dehydrogenase (3-fold); • Carnitine-Acylcarnitine carrier protein (3-fold); • Acyl CoA dehydrogenase ( 5-fold); • Isocitrate dehydrogenase (only TNF); • ADP/ATP translocase 2 ( 7.6-fold); • Cytochrome C1 ( 2-fold); • Electron transfer flavoprotein ( 2-fold); • VDAC-1 ( 2-fold); • Cytochrome C1 ( 2-fold); • Cytochrome C oxidase (TNF only); • Glycerol-3-phosphate dehydrogenase (TNF only) DAVID Bioinformatics Resources® UP REGULATED AND TNF-α ONLY: 13 proteins DAVID Bioinformatics Resources® UP REGULATED AND TNF-α ONLY: Function, pathway, process P-value Nucleic Acid Metabolism 0.000004 Protein Synthesis 0.00006 Protein Trafficking 0.00009 EIF-2 Signalling 0.000015 Glutathione Metabolism 0.0004 Interleukin Signalling 0.0007 Oxidative Stress 4x10-7 Mitochondrial Dysfunction 0.008 Ingenuity® Systems DOWN REGULATED AND CONTROL ONLY: 27 proteins CYTOSKELETON PROTEINS: • ADP Ribosylation Factor ( 5-fold); • Actin, alpha-1 ( 4-fold); • Actin, gamma-1 (control only); • Alpha actinin-4 ( 4-fold); • Cofilin-1 (5.5-fold); • Gelsolin ( 43-fold); • Tubulin, beta 2 ( 31-fold); • R-ras ( 52-fold) DAVID Bioinformatics Resources® DOWN REGULATED AND CONTROL ONLY: Function, pathway, process P-value Cellular Assembly & Organisation 1.7 x 10-9 Cellular Function & Maintenance 1.7 x 10-9 Cell Morphology 1.7 x 10-7 Cellular Growth & Proliferation 3.8 x 10-7 Integrin Signalling 2.4 x 10-10 Caveolar-mediated Endocytosis 4.5 x 10-9 Clathrin-mediated Endocytosis 3.3 x 10-8 Actin Cytoskeleton Signalling 2.7 x 10-7 Ingenuity® Systems EVIDENCE OF TNF-α SIGNALLING: Proteomics: • TNF-R1 and Death Associated Protein (TRADD): Expressed only in TNF-α stimulated cells • NF-κB: Expressed only in TNF-α stimulated cells Control 0.5ng/ml 5ng/ml TRADD 20ng/ml IκB-α β-tubulin Inflammatory / Immune Protein Expression: IκB EXPRESSION • Complement C4 (2.2-fold); • ICAM-1 (only TNF-α); • MHC Class 1 (1.6-fold); • IL-1 (TNF-α only) eNOS-NO Pathway: • • • • • eNOS: 27% eNOS: 63% NO: 44% NO: 33% NO: 23% NOS-NO BIOSYNTHESIS PATHWAYS: NO PRODUCTION Control TNF 0.5ng TNF 0.5ng TNF 20ng HSP90 β- tubulin Control TNF-α 5ng/ml p-PKB HEAT SHOCK PROTEIN 90 EXPRESSION t-PKB PKB/Akt REGULATION AND ACTIVATION PROTEOMICS: • Heat shock protein 90-α (5.8-fold) • Heat shock protein 90-β (42-fold) Oxidative Stress: • • • • ROS: 63% of studies Nitrosative stress: 25% of studies NADPH-oxidase: 25% of studies ROS included: Superoxide (50%), mito- ROS (25%) and H2O2 (13%) PROTEOMIC DATA SUGGEST ANTI-OXIDANT PROTEINS AND OXIDATVE STRESS RESPONSE: PROTEOMICS: • Park-7 ( 2-fold ) • SOD [Mn], mitochondrial (2-fold) • Thioredoxin ( 3-fold) • Glutathione-s-transferase (only in TNF) • Glutathione peroxidase, GPX4 (only TNF) DAVID Bioinformatics Resources® • Peroxiredoxin (2-fold) Function, pathway, process P-value Glutathione Metabolism 0.0004 Oxidative Stress 4x10-7 Ingenuity® Systems OXIDATIVE STRESS PARAMETERS: Control 0.5ng/ml 5ng/ml 20ng/ml p22-phox β-tubulin P22-PHOX EXPRESSION MITOCHONDRIAL ROS PRODUCTION: 20 µm Control: + MitoSoxTM 20 µm TNF-α: + MitoSoxTM FACS confirmed results Thus proteomic results confirmed Apoptosis / Cell Death: • Apoptosis: 50% of studies • Apoptosis: 38% of studies • Necrosis: 13% of studies APOPTOSIS / CELL DEATH: PROTEOMICS: • Bid (TNF only) • RACK-1 (2.7-fold) • PEA-15 (inhibits TNF-R1-mediated Caspase 8 activity) (6.3-fold) • VDAC-1 (1.6-fold) • BOK (TNF only) • Metadherin (anti-apoptotic) (TNF only) • Gelsolin (anti-apoptotic) ( 43-fold) Take home message???? • Vascular endothelial cells neglected in proteomics • CMEC basically no thorough study • This is novel and important study to gain knowledge into cardiovascular disease • Increase in oxidative stress due to TNF-α – proteins increase to counteract • eNOS – decreased HSP90 • Cells undergo apoptosis – increase in apoptotic proteins and increase in some anti-apoptotic proteins – therefore cells are fighting back • Due to increase in apoptotic proteins and hence increase in cell death the proteins involved in may be the result of cytoskeleton organisation which is decreased. Quantitative proteomic analysis of HIV-1 Tat apoptosis in SH-SY5Y neuroblastoma cells Putuma P. Gqamana1ǂ, Tariq Ganief1ǂ, Salome Smit2, Shaun Garnett1, Andrew Nel1, and Jonathan Blackburn1┴. Quantitative proteomic analysis of HIV-1 Tat induced apoptosis in SH-SY5Y neuroblastoma cells. Manuscript in preparation. • Tat associated with neural cell death and probable agent of HIV associated dementia • 2849 proteins were identified from SILAC treated cells which were either phosphoenriched or phospho-depleted (therefore reduced complexity of sample) • 17 up regulated and 72 down regulated proteins identified from SILAC • Dysregulation of proteins identified associated with several neurodegenerative disorders • Cell adhesion proteins down regulated – associated with apoptosis • Proteins identified may also have role in weakening of immune response • From results: • Tat neurotoxicity may activate early signalling via tyrosine phosphorylation receptors and cause mitochondrial and oxidative stress leading to apoptosis. This will form basis of future biomarker discovery for HIV associated dementia Putuma P. Gqamana1ǂ, Tariq Ganief1ǂ, Salome Smit2, Shaun Garnett1, Andrew Nel1, and Jonathan Blackburn1┴. Quantitative proteomic analysis of HIV-1 Tat induced apoptosis in SH-SY5Y neuroblastoma cells. Manuscript in preparation. Recent successes with Orbitrap Orbitrap Velos MS Neuroblastoma cells 7539 M. smegmatis 3271 M. Bovis 2368 P. falciparum 1681 CMEC 1663 V. Cholera 1411 Urine biomarkers 1500 Sample from another MS unit Single human peptide in Arabidopsis 546 vs 24 28 Acknowledgements Thanks to: Prof Hans Strijdom – US – CMEC And students Amanda, Mashudu and Corli Dr Putuma Gqamana – UCT – SILAC proteomics, neuroblastoma cells Dr Brandy Gqamana-Young – UCT – Urine proteomics Mae Newton-Foot and Zhou Fang – US – M. Smegmatis Louise Vos – US – M. Bovis Dr Martella du Preez and Lisa Schaeffer – CSIR – V. Cholera Dr Cobus Zwiegelaar – Azargen – human peptide 29 Proteomics Laboratory Senior Analyst: Dr Salome Smit Office: 021 938 9632 Fax nr : 086 690 7602 email: salomiesmit@sun.ac.za Universiteit van Stellenbosch Besoek / Visit: www.sun.ac.za/saf www.facebook.com/pages/CAF-Proteomics-lab-University-ofStellenbosch/278646975539969