Document
advertisement

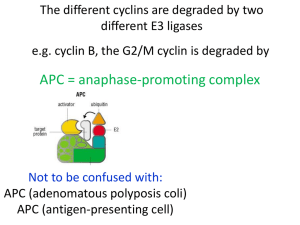
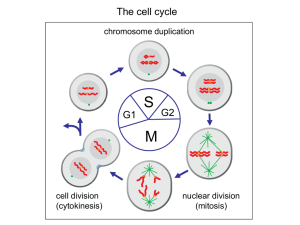
Regulation of Cytokinesis Corinna Benz, PhD, Biology Centre Overview • Cell cycle and cytokinesis in mammalian cells • Cell cycle and cytokinesis in trypanosomes • NDR kinase – MOB signalling in different organisms General idea The eukaryotic cell cycle Alberts et al., Molecular Biology of the Cell, 4th edition Different microtubules in a mammalian spindle Alberts et al., Molecular Biology of the Cell, 4th edition A condensed chromosome with attached kinetochore microtubules Alberts et al., Molecular Biology of the Cell, 4th edition Cell cycle control system • • • • Events must occur in a timely fashion Correct order Only once per cell cycle Backup system if things go wrong (checkpoints) • Adaptability Cell cycle checkpoints Alberts et al., Molecular Biology of the Cell, 4th edition The major players – Cyclindependent protein kinases (Cdks) • Oscillation between active and inactive states cyclical phosphorylation of cellular proteins that initiate or regulate cell cycle events • Activity regulated by binding to a cyclin partner (with exceptions) • Cyclins are degraded cyclically while Cdks are constitutively present Cyclin-dependent protein kinases (Cdks) and cyclins M-Cdk promotes mitotic events G1-Cdk promotes passage through a restriction point G1/S-Cdk commits cell to DNA replication S-Cdk is required for initiation of DNA replication Alberts et al., Molecular Biology of the Cell, 4th edition The major cyclins and Cdks of vertebrates and budding yeast Vertebrates Budding Yeast Cyclin-Cdk complex Cyclin Cdk partner Cyclin Cdk partner G1-Cdk cyclin D* Cdk4, Cdk6 Cln3 Cdk1** G1/S-Cdk cyclin E Cdk2 Cln1,2 Cdk1 S-Cdk cyclin A Cdk2 Cln5,6 Cdk1 M-Cdk cyclin B Cdk1** Clb1,2,3,4 Cdk1 * There are three D cyclins in mammals (cyclins D1, D2, and D3). ** The original name of Cdk1 was Cdc2 in both vertebrates and fission yeast, and Cdc28 in budding yeast Adapted from Alberts et al., Molecular Biology of the Cell, 4th edition Regulation via phosphorylation • Cdk-cyclin complexes are regulated through phosphorylation • Particularly important at onset of mitosis • Further regulation through Cdk inhibitor proteins (CKIs) Alberts et al., Molecular Biology of the Cell, 4th edition Control through proteolysis • SCF (Skp1-Cullin-F-box protein): Ubiquitin ligase, active in G1/S, constitutively active, substrates become available through phosphorylation • APC/C (anaphase-promoting complex): Ubiquitin ligase, active in G1 (subunit Cdh1) and M (subunit Cdc20), activated by addition of subunits APC/C complex SCF complex Control through proteolysis Alberts et al., Molecular Biology of the Cell, 4th edition The specifics Exit from G0 and G1 events Cdk7 Rb + growth factor stimulus cyclin D ↑ G0 to G1 transition E2f Cdk4:cyclin D/Cdk6:cyclin D G1-Cdk Rb- P negative feedback loop E2f transcription of e.g. cyclin A cyclin B cyclin E cyclin A/B cyclin E ↑ APC/CCdh1 cyclin A/B-ubiquitin Cdk2:cyclin E G1/S-Cdk G1 S degradation Exit from G0 and G1 events • Most cells in adults are in quiescent G0 stage • Growth factor stimulus cyclin D levels increase and Cdk4:cyclin D/Cdk6:cyclin D (G1-Cdk) drives cells into G1 by phosphorylating retinoblastoma tumor suppressor protein (Rb) • Phosphorylated Rb can no longer inhibit transcription factor E2f expression of genes important for DNA replication is induced • These genes include cyclin A, B and E: Only cyclin E accumulates in G1 since cyclin A and B are marked for degradation by APC/CCdh1 • Cdk2:cyclin E (G1/S-Cdk) promotes G1-S transition Genome duplication: Replication licensing 1) Helicase loader Mcm helicase (inactive) DePamphilis et al, Frontiers in PHYSIOLOGY, 2012 Genome duplication • Initiated at replication origin, where a preRC (prereplication complex) is assembled which is then converted to a preIC (pre-initiation complex) • Replication licensing = Assembly of preRCs on chromatin • Replisome: DNA replication proteins • preRC assembly occurs only when Cdk activity is low (anaphase to G1/S transition) • 1) Assembly of helicase loader: Orc1-6, Cdc6 and Cdt1 • 2) Assembly of Mcm helicase (inactive): Mcm2-7 (double hexamer) Replication initiation 1) 2) DePamphilis et al, Frontiers in PHYSIOLOGY, 2012 Replication initiation 3) 4) DePamphilis et al, Frontiers in PHYSIOLOGY, 2012 Replication initiation 5) DePamphilis et al, Frontiers in PHYSIOLOGY, 2012 Replication initiation • DDK (Dbf4-dependent Cdc7 kinase) phosphorylates Mcm4 and 6 • Sld3 and Cdc45 assemble into preRC • Sld2 and 3 are phosphorylated by Cdk2:cyclin E (G1/SCdk) • Sld2-P and Sld3-P bind to Dpb11 recruitment of GINS complex • Recruitment of Pol-ε (leading strand synthesis) • ssDNA binding protein RPA required to facilitate DNA unwinding • Formation of two active replisomes upon addition of Polα:primase and Pol-δ • Mcm10 stabilises final replisome structure DNA replication checkpoints 2) 3) DePamphilis et al, Frontiers in PHYSIOLOGY, 2012 DNA replication checkpoints • Effector kinase: Chk1 • Inhibits activation of preRCs by: • Phosphorylating and inhibiting phosphatase Cdc25, which then prevents activation of Cdk2 • Phosphorylating Sld3 (at a site different from the one phosphorylated by Cdk2:cyclin E (G1/S-Cdk)), which prevents interaction with Dpb11 (prevents formation of Cdc45:Mcm:GINS complex) Prevention of DNA re-replication DePamphilis et al, Frontiers in PHYSIOLOGY, 2012 Prevention of DNA re-replication • Cdk2:cyclin A (S-Cdk) phosphorylates Orc1, Cdc6 and Cdt1 export to cytoplasm and/or degradation • Absence of Orc1 causes other Orc subunits to leave the complex • Non-phosphorylated Cdt1 bound to PCNA (polymerase processivity factor) is ubiquitinated by CRL4:Cdt2 • Re-expression of preRC proteins in G2 and M phase, but in modified forms which are kept inactive • Metaphase exit: Cdc14 de-phosphorylates Orc subunits and Cdc6 allowing preRC assembly • Geminin degraded Cdt1 becomes available for preRC assembly DNA damage response Wang et al., Molecular Cancer, 2009 DNA damage response • Checkpoint kinases Chk1 and Chk2, activated by ATR and ATM kinases • ATM activated by ionising radiation • Chk2 in ATM-Chk2-Cdc25 pathway that senses double strand breaks, not essential in mammals • ATR activated by UV radiation • Chk1 in Atr-Chk1-Cdc25 pathway that senses single strand breaks, bulky lesions and stalled replication forks, essential in mammals • Both pathways result in Cdc25 phosphorylation sequestered in cytoplasm by 14-3-3 M-Cdk inactive • p53 phosphorylation causes activation of p21 (M-Cdk inhibitor) and upregulation of 14-3-3 Exit from S phase CRL1:Skp2 P cyclin E-P Cdk2:cyclin A S-Cdk E2f-P P P Cdk1:cyclin B M-Cdk Cdk1:cyclin B-P cyclin E-P -ubiquitin DNA replication proteins ↓ preRC proteins ↓ translocates to nucleus phosphorylates substrates to initiate mitosis Exit from S phase • Cyclin E inactivated through phosphorylation by Cdk2:cyclin A (S-Cdk) becomes a target for CRL1:Skp2 ubiquitin ligase and is degraded • Cdk2:cyclin A (S-Cdk) also phosphorylates and inactivates E2f (downregulation of expression of genes required for DNA replication, also prevents re-licensing of origins) • Cyclin B levels increase Cdk1:cyclin B (M-Cdk) complex formed, translocates to nucleus to phosphorylate substrates important for entry into mitosis Entry into mitosis – M-Cdk activation P P Wee1 Y15-P Myt1 Cdc25 Cdk1 Cdk1 Cdk1:cyclin B M-Cdk Y15-P T14-P T14-P G1, S, G2 M G1 Positive feedback loop Entry into mitosis – APC/C activation S/G2: Emi1 APC subunits: * Cdc20-P active * Cdh1-P inactive Cdc20-P Cdk2:cyclin A S-Cdk P APC/C Cdh1 binding degradation Prometaphase: Emi1 APC/CCdc20 Cdk1:cyclin B M-Cdk P Cdc20-P APC/CCdc20 Cdk2 Entry into mitosis cyclin A p21/p27 cyclin B Cdk1 Anaphase progression Entry into mitosis • • • • • Positive feedback loop increases activity of Cdk1:cyclinB (M-Cdk) APC/C regulation: Cdc20 subunit is active when phosphorylated Cdh1 subunit is inactive when phosphorylated During S and G2: APC/C inactive, Emi1 prevents phosphorylation of Cdc20 and Cdk2:cyclin A (S-Cdk) phosphorylates the APC/C, which inhibits binding of Cdh1 and results in its degradation • Prometaphase: Selective Emi1 degradation • Negative feedback loops regulate Cdk1/2 activity through APC/Cmediated ubiqitination of cyclin A and B • Cdk1 and Cdk2 also regulated by interaction with Cdk inhibitors (CKIs) p21 and p17 whose expression follows that of the APC/C M-Cdk – functions • Induces assembly of mitotic spindle • Ensures that replicated chromosomes attach to spindle • In many organisms also triggers chromosome condensation, nuclear envelope breakdown, actin cytoskeleton rearrangement, reorganisation of Golgi and ER; e.g. through phosphorylation of lamins dismantling of nuclear envelope Metaphase-anaphase transition Alberts et al., Molecular Biology of the Cell, 4th edition Metaphase-anaphase transition • M-Cdk phosphorylates condensin complex resulting in chromosome condensation at prometaphase • M-Cdk activates APC/CCdc20 triggers anaphase by promoting degradation of securin • Securin no longer inhibits the protease separase • Separase becomes active and cleaves cohesin subunits resulting in sister chromatid separation Spindle attachment checkpoint • Mitotic spindle in green • Mad2 in red • Sister chromatids only separated when all chromosomes properly attached to spindle • State of kinetochore monitored • Several proteins e.g. Mad2 recruited to unattached kinetochores inhibition of APC/CCdc20 Alberts et al., Molecular Biology of the Cell, 4th edition Dai and Grant, Clinical Cancer Research, 2010 Cytokinesis Agromayor and Martin-Serrano, TRENDS in Cell Biology, 2013 Cytokinesis initiation • Signalling between anaphase spindle and cortex • Spindle recruits narrow zone of active RhoA (GTPase) • Active RhoA recruits effector contractile ring proteins (cytokinesis formin, Rho kinase, Citron kinase) • RhoA flux model: Global GAP-mediated RhoA inhibition versus localised GEF-mediated RhoA activation (e.g. Ect2 at cell equator) GAP=GTPase activating protein, GEF=Guanine nucleotide exchange factor Green et al, Annu. Rev. Cell Dev. Biol., 2012 Central spindle (spindle midzone) formation • Spindle midzone: Overlapping, antiparallel microtubules (MTs) (+ ends facing each other) • Formation requires PRC1 and kinesins Kif4 and MKLP1 • PRC1: antiparallel MT cross linker • Chromosomal Passenger Complex (CPC): Aurora B (kinase) and three additional subunits phosphorylates and recruits • Centralspindlin: Heterotetramer, consists of two molecules MKLP1 and two molecules CYK-4 (GAP) Green et al, Annu. Rev. Cell Dev. Biol., 2012 Central spindle (spindle midzone) formation • PRC1 recruits Kif4 to overlap zones where it slows down MT dynamics • Polo-like kinase 1 (Plk1) required for spindle elongation • Plk1 recruits itself by phosphorylating substrates (e.g. PRC1, MKLP2) and thus creating binding sites • Plk1 phosphorylates CYK-4 subunit of centralspindlin, which then scaffolds recruitment of Ect2 Green et al, Annu. Rev. Cell Dev. Biol., 2012 Central spindle (spindle midzone) formation • Ect2 (GEF for RhoA) converts RhoAGDP into RhoA-GTP, which triggers contractile ring assembly • Contractile ring consists of actin, myosin II and septin filaments (recruited by anillin which as a crosslinker binds to all three) • Ring is disassembled as it constricts Green et al, Annu. Rev. Cell Dev. Biol., 2012 Spindle midzone midbody • Upon contractile ring constriction, midzone-localised proteins are redistributed: • PRC1 and Kif4 stay at MT overlap zone • Centralspindlin and Ect2 concentrate in midbody ring, where they colocalise with anillin, RhoA, ARF6 and Cep55 • CENP-E, MKLP2 and Aurora B colocalise with tightly packed, parallel midbody MTs in regions flanking the midbody core • Plk1 essential for these relocalisations Green et al, Annu. Rev. Cell Dev. Biol., 2012 Contractile ring midbody ring • Anillin required for assembly of midbody ring and anchoring to plasma membrane • Citron kinase essential for abscission, required for localisation of anillin and RhoA • RhoA required for anillin localisation to midbody ring Green et al, Annu. Rev. Cell Dev. Biol., 2012 Abscission Agromayor and Martin-Serrano, TRENDS in Cell Biology, 2013 Abscission • CEP55, TSG101 and ALIX are translocated to midbody and mediate recruitment and polymerisation of ESCRT-III (complex for scission) • MIT-domain containing protein 1 (MITD1) coordinates activity of ESCRT-III • Rab35 recruits OCRL and Rab11/FIP3-positive endosomes deliver p50RhoGAP to midbody resulting in changes of membrane lipid composition and clearing of actin • FYVE Cent interacts with PtdInsP and recruits TTC19 and CHMP4B • ESCRT-III filaments constrict midbody • Spastin (MT severing enyzme) cleaves microtubules • AAA ATPase VPS4 disassembles ESCRT-III Abscission checkpoint Agromayor and Martin-Serrano, TRENDS in Cell Biology, 2013 Abscission checkpoint • CHMP4C (ESCRT-III subunit) regulates Aurora Bmediated abscission checkpoint • Trapped chromatin at midbody CHMP4C binds to Borealin and is phosphorylated by Aurora B • CHMP4C-P relocalises to midbody preventing abscission until chromatin is removed The cell cycle and cytokinesis in Trypanosoma brucei • Several single copy organelles/structures (e.g. flagellum) that need duplicated and segregated • Cell division achieved by lateral ingression of a cleavage furrow rather than medial constriction of actin filaments • Actin is dispensable for cytokinesis Wheeler et al, Molecular Microbiology, 2013 Cdk-cyclin system in T. brucei • Cdk-cyclin system is conserved • 10 cyclins and 11 CRKs (Cdc2-related kinases) • 26 potential interactions between these have been identified • Not all of them exclusively involved in cell cycle regulation (CRK12, CYC2 and CYC7) Ziyin Li, Eukaryotic Cell, 2012 DNA replication and licensing • Orc components: Orc1/Cdc6 related protein, Orc1b, Orc4, Tb3120 and Tb7980 • Well-conserved CMG (Cdc45-Mcm-GINS) complex, but homologs of Cdt1, Sld2, Sld3 and Cdc7-Dbf4 missing • Additional licensing system for mitochondrial DNA (kDNA) Yeast T. brucei Ziyin Li, Eukaryotic Cell, 2012 T. brucei mitosis • Closed mitosis (no nuclear envelope break down) • MTOCs (flagellar basal bodies) not involved in spindle formation • Mitotic kinesin Kif13-1 • APC/C components expressed, only APC1 and Cdc27 essential Ziyin Li, Eukaryotic Cell, 2012 for mitosis, substrates? • Aurora B (AUK1) forms unique CPC with two novel proteins • Conserved proteins like INCENP, Survivin and Borealin absent Differential localisation of trypanosome CPC during mitosis • Trypanosome CPC shows dynamic localisation: • Chromosomes to central spindle • Central spindle to anterior tip of the new flagellar attachment zone (FAZ) • Travels along cleavage furrow to the posterior end of the cell Li et al, PLoS ONE, 2008 Cytokinesis signalling in trypanosomes vs metazoa • AUK1 and PLK implicated in cytokinesis • Both proteins found at anterior tip of new FAZ during late stages of cell cycle • Common target? • Rho-like small GTPase, RHP present and involved in cytokinesis, but doesn’t localise to cleavage furrow • Role of MOB1, PK50, PK53 and Cdc14? Ziyin Li, Eukaryotic Cell, 2012 A closer look at Mob-NDR signalling S. cerevisiae D. melanogaster H. sapiens yellow: STE20-like kinase green: NDR kinase red: MOB protein Hergovich, Cellular Signalling, 2011 Common Elements of Regulation • STE20-like kinases phosphorylate MOB proteins • Phosphorylated MOB proteins can interact with and activate NDR kinases • NDR kinases phosphorylate downstream targets and function in mitotic exit and morphogenesis in yeast, morphogenesis and cell proliferation in Drosophila and centrosome duplication and cell proliferation in mammalian cells FEAR and MEN in budding yeast Bub2-Bub1: GAP Tem1: GTPase Lte1: GEF D'Amours and Amon, Genes and Development, 2004 • Cdc14 is needed for mitotic exit in budding yeast • Cdc14 is kept inactive throughout the cell cycle by binding to Net1 • Polo kinase (Plk) activates Cdc14 in early anaphase (FEAR = Cdc fourteen early anaphase release) • Cdc14 dephosphorylates/activates Cdc15 (STE20-like kinase) • Cdc15 phosphorylates Dbf2-Mob1 and activates the complex • Dbf2-Mob1 phosphorylates Cdc14 and activates it Role of PLK, NDR kinases, MOB1 and Cdc14 in T. brucei PLK (polo) RNAi PK50 RNAi post-mitotic cells that are not dividing MOB1 RNAi PK53 RNAi • PLK, the NDR kinases PK50 and PK53, MOB1 and Cdc14 are essential in trypanosomes • Depletion results in specific cytokinesis defects Cdc14 RNAi post-mitotic cells that are arrested during cytokinetic furrow ingression Hammarton et al, Molecular Microbiology, 2005; Hammarton et al, Molecular Microbiology, 2007; Benz, Ma et al, JBC, 2010 Differential regulation of trypanosome NDR kinases Recombinant trypanosome NDR kinases are active Trypanosome NDR kinases do not interact with MOB1 proteins Benz, Ma et al, JBC, 2010 Differential regulation of trypanosome NDR kinases • Recombinant trypanosome NDR kinases are active and not further activated in the presence of MOB proteins • Endogenous trypanosome kinases don’t interact with MOB proteins • PLK is excluded from the nucleus during the entire cell cycle and does not function in mitosis • NDR kinases, MOB proteins and Cdc14 are not substrates of each other • Regulation and signalling pathways??? • Conserved proteins ≠ conserved functions! Questions?