BCHM 300 Introduction to Structural Biology (2011) lecture 1
advertisement

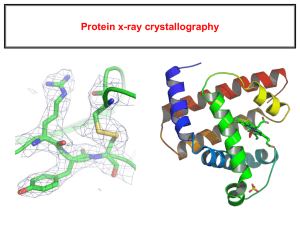
Jan. 5, 2011 Biochemistry 300 Introduction to Structural Biology Walter Chazin 5140 BIOSCI/MRBIII E-mail: Walter.Chazin@vanderbilt.edu http://structbio.vanderbilt.edu/chazin/classnotes/ Structural Biology- Multiple Scales 3D structure R - N - Ca- CO H polymerase SSBs Atoms Complexes helicase primase Organism Assemblies Cell Structures Cell The Underlying Basis for Biology Organ Tissue Cell Molecule Atoms • A cell is an organization of millions of molecules • Proper communication between molecules is essential to normal functioning of cells and miscommunication is the basis for disease • To understand the basis for communication it is necessary to define the atomic structures of the molecules and to elucidate the fundamental forces driving interactions between them Atomic Resolution Structural Biology Determine atomic structure to analyze why molecules interact The Reward: UnderstandingControl Anti-tumor activity Duocarmycin SA Atomic interactions Shape Atomic Structure in Context NER RPA BER RR Molecule Pathway Activity Structural Genomics Structural Proteomics Struct. Systems Biol. See commentary by SC Harrison, NSMB 11, 12-15 (2004) Techniques for Atomic Resolution Structural Biology NMR Spectroscopy X-ray Crystallography Computation Determine experimentally or model 3D structures of biomolecules Structures from X-ray Crystallography and NMR are Generated Differently X-ray X-rays Diffraction Pattern NMR RF Resonance RF H0 Direct detection of atom positions Crystals Indirect detection via H-H distances In solution Why Compute Structures? • Crystallography and NMR don’t always work! – Many important proteins do not crystallize – Size limitations with NMR • A good guess is better than nothing! – Enables the design of experiments – Potential for high-throughput • Invaluable for analyzing/understanding structure Computational Approaches Molecular Simulations • Convert experimental data into structures • Predict effects of mutations, changes in environment • Insight into molecular motions • Interpret structures- characterize the chemical properties (e.g. surface) to infer function Computational Approaches Structure Prediction 1 QQYTA KIKGR 11 TFRNE KELRD 21 FIEKF KGR Algorithm • Secondary structure (only sequence) • Homology modeling (using related structure) • Fold recognition • Ab-initio 3D prediction: “The Holy Grail” Complementarity of Methods • X-ray crystallography- highest resolution structures; faster than NMR • NMR- in solution; enables widely varying conditions; can characterize dynamic, weakly interacting systems and movement • Computation- models without experiment; very fast; fundamental understanding of structure, dynamics and interactions; provides insight into driving forces There is No Such Thing as A Structure! • Polypeptides are dynamic and therefore occupy more than one conformation- Structural Dynamics Is there a specific biologically relevant conformer? Does a molecule crystallize in a biologically relevant conformation? What about proteins and protein machines with architecture that is not fixed? Molecules are Dynamic, Not Static Conformational Ensemble “Neither crystal nor solution structures can be properly represented by a single conformation” Intrinsic motions Imperfect data Variability reflected in the RMSD of the ensemble Representing Molecular Structure C N A representative conformer from the ensemble How is Motion Reflected in X-ray Crystallography and NMR? X-ray NMR • Uncertainty Avg. Coord. + B factor Ensemble Coord. Avg. • Flexibility Diffuse to 0 density Multiple occupancy Mix static + dynamic Sharp signals Fewer interactions Measure motion! Challenges For Understanding The Meaning of Structure • Structures determined by NMR, computation, and X-ray crystallography are static snapshots of highly dynamic molecular systems • Biological process (recognition, interaction, chemistry) require molecular motions (from femto-seconds to minutes) • New methods are needed to comprehend and facilitate thinking about the dynamic structure of molecules: visualize structural dynamics Visualization of Structures Intestinal Ca2+-binding protein! Need to incorporate 3D and motion Addressing Complex Systems: The Divide and Conquer Strategy • Cellular machinery has large and complicated structures not readily amenable to high resolution techniques • Characterize the stable folded domains at the atomic level and elucidate driving forces • Build up a structural model of the whole from a reconstruction with the high resolution pieces Validate by experiments on the intact protein(s) and functional analysis Need Additional Techniques For Large Molecules/Complexes NMR Spectroscopy X-ray Crystallography Computation Determine experimentally or model 3D structures of biomolecules • EPR/Fluorescence to measure distances when traditional methods fail • EM and Scattering to get snapshots of whole molecular structures (Cryo-EM starts to approach atomic resolution!) Snapshots of Molecular Assemblies Very large structures lower resolution MBP-tagged Siah-1 Stewart Lab Inserting High Resolution Structures into Low Resolution Envelopes Mesh = DAMMIN Ribbon = 1QUQ The Horizon: Dynamic Protein Machinery Activity Requires Remodeling of Multi-Protein Assemblies Thinking in Terms of Protein Architecture 14/32D/70C 70AB X-ray Zn P C D 14 32CTD B A RPA70 RPA32 CTD RPA14 NTD quaternary structure? NMR 70NTD Dynamic Architecture of Proteins in Molecular Machines Movement/remodeling of architecture is intrinsic to function!! Center for Structural Biology Dedicated to furthering biomedical research and education involving 3D structures at or near atomic resolution http://structbio.vanderbilt.edu