SCSB Resources Page August 2013 - Sealy Center for Structural
advertisement

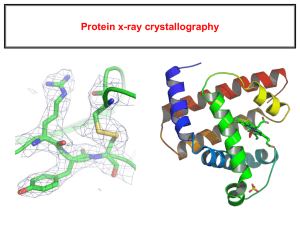
UTMB Sealy Center for Structural Biology and Molecular Biophysics (SCSB): The SCSB comprises five instrumentation and resource centers: NMR, X-ray Crystallography, Solution Biophysics, Computational Biology, and Cryo-Electron Microscopy, described below. The center provides instrumentation time courtesy of a center endowment making them free to center researchers. All resources described below are located on the University of Texas Medical Branch at Galveston campus, located approximately 50 miles from the main Texas Medical Center in Houston. NMR Spectroscopy Laboratory: Solution structure and dynamics measurements of biological macromolecules as well as small molecules are facilitated through the NMR Laboratory. Resources are located in a dedicated two-story NMR building on the UTMB campus with a combined area of over 9,000 ft2. Adjoining the NMR instrumentation lab is a wetlab for sample preparation. The current instrumentation includes the state-of-the-art high field Bruker Avance III 800MHz (with TCI cryoprobe), 750MHz, and 600 MHz (with QCI cryoprobe) NMR spectrometers, which are capable of various multi-dimensional 1H/2H/13C/15N/31P NMR experiments. These instruments include new Bruker AVANCE™ III 800, 750 AND 600 MHz consoles installed in 2011, along with new probes. The research of the NMR laboratory is focusing on NMR methods, structure determination, protein dynamics and function, small molecules and metabolomics study. The core research laboratory offers instrumentation, training, software and assistance in designing NMR experiments. X-ray Crystallography Laboratory: This laboratory provides complete facilities for crystallization studies of macromolecules. The facility houses a Rigaku Crystalmation ® robotics system, consisting of an Alchemist-II for preparation of crystal screens, a PHOENIX, for fast reliable and efficient screening of crystallization conditions, and a DT Minstrel/Gallery 160 for automated imaging of crystallization trays. Two X-ray area detector systems are available for the collection of X-ray diffraction data: The Rigaku Ultimate Homelab, comprising an FR-E++DW superbright X-ray generator and R-AXIS++VI crystallography system, provides high resolution data and rapid screening of samples, in addition to in-house phasing using chromium radiation. The Bruker 2K CCD provides fast screening and small molecule structure solution capabilities. These are suitable for protein crystals from 20 microns in size with cell dimensions up to 400 Å and are each equipped with cryogenic sample cooling systems. Solution studies are performed using our Malvern µV DLS system and Rigaku BioSAXS-1000. The fully equipped Rigaku BioSAXS-1000 Kratky SAXS camera coupled with the FR-E++ X-ray source provides the best home-lab SAXS camera currently available for biological samples. UTMB is also one of just a select few institutions to have in-house Biological SAX capabilities, and was the first in the country to use BioSAX on an FR-E++ system. The quality of the data can approach that from the synchrotron. The camera produces usable data from q~0.01 Å-1 to q=0.6 Å-1. The Crystallographic Computation Lab provides high-performance Linux stereographics workstations for crystallographic structure solution, refinement, and analysis or CryoEM reconstructions. Diffractometer, SAXS camera, and robotics scheduling, for qualified users, is available online. The X-ray crystallography laboratory is staffed with 1.5 FTE PhD level scientists who can provide training, experimental design, data processing, and structure determination to users as needed. The X-ray Crystallography Laboratory is also a member of the Gulf Coast Protein Crystallography Consortium line at the CAMD PX Beamline. The GCPCC beamline is optimized for SAD/MAD data collection. It features a newly installed 7.5T multipole wiggler coupled to either a channel-cut or a multilayer monochromator both of which provide a tunable high-intensity x-ray source for the Marresearch CCD endstation. Cryo-Electron Microscopy (cryo-EM) Laboratory: The Laboratory (including the W. M. Keck Virus Imaging Center) has been completely rebuilt including new instrumentation as a result of the 2008 Hurricane Ike storm surge. The recently renovated Laboratory houses two state-of-the-art 200 keV cryo-electron microscopes, a JEM 2200FS and a JEM 2100, with remote imaging capabilities that allow microbiologists and other researchers worldwide to directly access the Laboratory for data collection and image processing. The JEM 2200FS is located in the W. M. Keck Virus Imaging Center, the first of its kind BSL-3 cryo-EM facility to provide the ability to work with live wild type infectious agents. A third, 100 keV, JEM 1400 microscope is available for both Cryo- and conventional EM studies including negatively stained samples and plastic thin sections of cells and tissues. It is used for sample screening, lower resolution studies and for user training. All the microscopes in the Laboratory have digital cameras (Gatan) for image acquisition and software for automated data collection. The Laboratory allows for advanced structural studies of macromolecular complexes and cellular organelles using different techniques, e.g. single particle imaging and electron tomography. While single particle imaging is used for studying structure of ensembles of identical macromolecules, electron tomography, including cryo-electron tomography, can be used to study samples with unique structure and individual events, e.g. virus-host cell interactions, viral entry into cells, structure of pleomorphic viruses, cell components, bacteria, etc. The W. M. Keck Virus Imaging Center promotes extensive and productive collaborations between virologists, microbiologists and electron microscopists to obtain and analyze high resolution structures of infectious agents to understand the mechanism of their virulence and infectivity; the whole Laboratory serves as an intellectual core drawing together and helping researchers in various fields at UTMB and around the country. The Cryo EM Laboratory houses two separate cryo-preparation rooms, an open research specimen preparation room with Vitrobot and manual cryo-plungers, and a BSL-3 specimen preparation room with a manual cryo-plunger; a Gatan Solarus plasma cleaner; a Cressington 308 vacuum coater; a Leica EMPACT2 high-pressure freezer; an AFS2 freeze-substitution system from the same company, and a Leica UC7 ultramicrotome furnished with FC7 cryo-chamber and other ancillary equipment. A new generation 4kx5k direct electron detection camera (DE-20) with dose fractionation (i.e. “movie”) capabilities was recently installed in the BSL-3 2200FS cryoEM. This state-of-the-art camera is capable of producing up to 40 frames/s image series, allowing for specimen drift compensation and radiation damage correction. The Laboratory has several workstations for image processing and shares a computing facility with the X-ray Crystallography Laboratory that includes 4 multi-processor Xeon/Opteron servers/ workstations and the 120 core EM computational cluster. The Cryo EM Laboratory has 1.5 FTE PhD Level scientists to assist in sample preparation, experiment design, data processing, training, and BSL-3 Cryo EM work. Solution Biophysics Laboratory: This facility is housed on the fifth floor of the Medical Research Building comprising 1,200 ft2. Major instrumentation in the laboratory include: 1) XL-A Analytical Ultracentrifuge (Beckman-Coulter) – Sedimentation Velocity and Equilibrium experiments recorded at a wide range of absorbance wavelengths can be performed with this instrument. This technique is especially well suited for the analysis of oligomerization and the evaluation of protein-protein interaction thermodynamics. (2) Voyager-DE STR MALDI-TOF Mass Spectrometer (PerSeptive Biosystems) - routinely used to check purity of protein and peptide samples. (3) Biacore T100 Surface Plasmon Resonance (GE Health Sciences) – the use of Surface Plasmon Resonance allows for the analysis of the kinetics and thermodynamics of protein-protein interactions. High-throughput capability is also available in this system. (4) ThermoFluor (Johnson & Johnson) – high throughput screening assay instrument. Thermal denaturation of samples in the presence of fluorescent dyes allows the analysis of the relative stability of proteins in the presence of different ligands. (5) Circular Dichroism Spectrometer J-815 (Jasco) – probes the structure of proteins in solution under varying conditions of pH, temperature, or other solutes. (6) Fluorometer Spex FluoroMax (Horiba) – fluorescence spectroscopy is widely used to obtain the thermodynamics of ligand binding and the stability of proteins. Quenching experiments can also be used to probe the degree of exposure of intrinsic fluorescent probes. (7) Isothermal Titration Calorimetry (Microcal/GE Health Sciences) – Used to obtain the thermodynamics of protein-ligand interactions. The Solution Biophysics Laboratory is managed by Dr. Luis Marcelo F. Holthauzen. Dr. Holthauzen trains new users, conducts more technical experiments and aids in experimental design and data analysis. Computational Biology & Bioinformatics The Sealy Center for Structural Biology and Molecular Biophysics provides access to researchers a number of computational resources. The SCSB is a partner in the UT System Research Cyber infrastructure (UTRC) initiative which connects UTMB to UT Austin's TACC. This connection is being upgraded to include a 10 Gbps UTNet intra-system connection with direct links to LEARN. The Lonestar Education and Research Network (LEARN) is a consortium of 38 public and private institutions throughout Texas. The LEARN network services include Connection Gateways to the National Lambda Rail and Internet2. The UTRC also provides 5 PB of storage in a UT Data Repository (UTDR) to enable long-term data storage, hosting of large scientific data collections for collaborations and communities, and analysis capabilities that span multiple collections. UTDR will facilitate data management proposals to NSF and NIH which is now a requirement in all NSF and NIH proposals. TACC operates two of the most powerful and capable high performance computing systems in the world; Ranger (579.4 teraflops and Lonestar 4 (302 teraflops) and soon Stampede (estimated at 10 petaflops) SCSB researchers are also users (millions of hours) of the San Diego Supercomputing facility including Triton (20 teraflops) Trestles (100 teraflops), Dash (4.9 teraflops) and Gordon (285teraflops) for large-memory analysis.