Structure Validation
advertisement

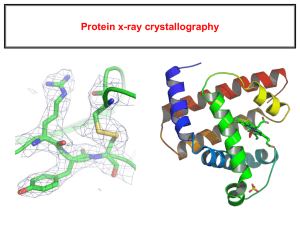
Protein structure validation Seminar series 2 Structure validation Everything that can go wrong, will go wrong, especially with things as complicated as protein structures. What is real? What is real? ATOM ATOM ATOM ATOM ATOM ATOM ATOM ATOM 1 2 3 4 5 6 7 8 N CA C O CB CG CD1 CD2 LEU LEU LEU LEU LEU LEU LEU LEU 1 1 1 1 1 1 1 1 -15.159 -14.294 -14.694 -14.350 -12.829 -11.745 -11.895 -10.378 11.595 10.672 9.210 8.577 10.836 10.348 11.027 10.636 27.068 26.323 26.499 27.502 26.772 25.834 24.495 26.402 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 18.46 9.92 12.20 13.43 13.48 15.93 13.12 15.12 X-ray X-ray X-ray ‘FFT-inv’ FFT-inv And now move the atoms around till the calculated reflections best match the observed ones. X-ray refinement / multiple minima Multiple minima X-ray R-factor Error = Σ w.(obs-calc) R-factor = Σ w.|obs-calc| 2 X-ray resolution NMR data collection From NMR spectra to structure B A If proton A and proton B are close in space, socalled ‘cross peaks appear’ in a spectrum due to the Nuclear Overhauser Effect (NOE). The NOE depends on the distance between proton A and B From NMR spectra to structure B A H1 A A B C .. H2 B C D D .. Distance(Å) 3 4 2 1 .. The NMR data thus contains distance information! Most NOEs are between close neighbours in the sequence. Those hold little information. The ‘good’ NOEs are between atoms far away in the sequence. There are few of those, normally. From NMR spectra to structure The list of distances can be used in a computer simulation that is reminiscent of protein folding. From NMR spectra to structure .. until the protein is ‘folded’. From NMR spectra to structure NMR ‘ensemble’ NMR refinement Multiple minima NMR refinement Green lines: Distance OK Red lines: Protons too far away from each other NMR Q-factor Error = Σ NOE-violations + Energy term2 NMR versus X-ray With X-ray you measure reflections. Each reflection holds information about each atom. With NMR you measure pair-wise distances, angles, and orientations. These all hold local information. X-ray requires crystals, and crystals cause/are artefacts. NMR is in solution, but provides much less precision. NMR versus X-ray ‘Error’ Mobility Crystal artefacts Material needed Cost of hardware Drug design NMR 1-2 Å yes no 20 mg 4 M Euro no X-ray 0.1-0.5 Å not really yes 1 mg near infinite (share) almost Better combine and use the best of both worlds. More about (protein) crystallography and NMR in: Kristalstructuur Magnetische Resonantie Fourier Analyse and Structuur, Functie, Bioinformatica, of course Why validation ? Why have we spend twenty years to search for millions of errors in the PDB? Validation because: Everything we know about proteins comes from PDB files. Errors become less dangerous when you know about them. And, going back to the connecting thread through this series, if a template is wrong the model will be wrong. What kind of errors can the software find? Administrative errors. Crystal-specific errors. NMR-specific errors. Really wrong things. Improbable things. Things worth looking at. Ad hoc things. Smile or cry? A B C D 5RXN 7GPB 1DLP 1BIW 1.2 2.9 3.3 2.5 Little things hurt big X-ray specific His, Asn, Gln ‘flips’ Hydrogen bond network Your best check: Contact Probability Contact Probability Contact probability box A positive nitrogen around a Phe X-ray How bad is bad? In a normal distribution, 68% of the points are within 1 standard deviation of the mean In a normal distribution, half of the points are above and half of the points are below average ΔG = -RT ln K 95% are within 2 sd Less than 1 in 10000 points is more than 4 sd away from the mean One slide about homology modelling How difficult can it be? 1CBQ 2.2 A How difficult can it be? How difficult can it be? How difficult can it be? How difficult can it be? 1CBQ 0.33 1.00 0.33 0.33 0.33 2.2 A How difficult can it be? This is what standard viewers show: 1CBQ 2.2 A Even if the oxygen labels would have been reversed, the so-called ‘asymmetric unit’ could also have been chosen in a much better way… Errors or discoveries? Buried histidine. Warning for buried histidine triggered biochemical follow -up and new mechanism for KH-module of Vigilin. (A. Pastore, 1VIG). Acknowledgements: Rob Hooft Robbie Joosten Elmar Krieger Sander Nabuurs Chris Spronk Maarten Hekkelman