- BUGS McGill
advertisement

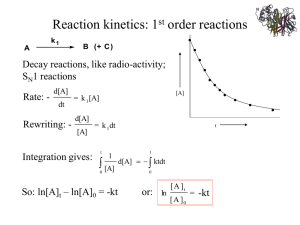
Lecture 2: Spectroscopy and Enzyme Kinetics By: Urshila Overview Spectroscopy Absorbance Beer-Lambert Law Sample Questions Enzyme Kinetics Rates and Michaelis-Menten Enzyme Inhibition Competitive vs. Non-competitive Inhibition constant Sample Questions Absorption Transmittance is the fraction of light passed through an object •Absorbance is measured using a spectrophotometer •It consists of 4 main parts: •Light Source •Monochromator (slit) •Sample (in a cuvette) •Light detector and recording device (usually a computer) Transmittance values range from 0 (all of the light is absorbed) to 1 (all of the light is transmitted) T = [I]/[I0] Absorbance is the amount of light absorbed by an object Absorbance is measured as the negative log of transmittance (T) Abs (or O.D) = -log10 (T) Beer- Lambert Law Absorbance is proportional to the A=ε*c*d concentration of a substance (c) and the distance that light travels through the substance (d = path length) This is denoted by the Beer-Lambert Law where: A= absorbance ε = molar extinction coefficient (mol-1 * L * cm-1 ) c= molar concentration (mol/L) d= path length (cm) Exceptions to the Beer-Lambert Complex Solutions Like Iodine, equilibrium between colored and non-colored species Nonhomogenous Solutions Scattering Solutions Particles shield each other from light at high concentration Particles can absorb, transmit, or scatter light Things to remember! Rules of thumb Protein: 1 O.D. @ 280 nm = 1 mg/ml DNA (ss) 1 O.D. @ 260 nm = 30 ug/ml DNA (ds) 1 O.D. @ 260 nm = 50 ug/ml Bacteria : 1 O.D. @ 600 nm = 5* 108 bacteria/ml SPF SPF = 1/T Doubling the thickness of sunscreen, doubles the absorbance and decreases transmittance by a factor of 100. (Recall: A = -log10 (T) ) Thinking Time 1. A researcher is studying the absorbance level of a bacterium at 600 nm and discovers that the absorbance spectrum does not follow the Beer-Lambert Law. What is a plausible explanation for this situation? 2. In what other situations would the Beer-Lambert Law not apply? 3. If you are measuring the spectra of a mixture, what modifications should be made to the Beer-Lambert Law? Answers 1. The large size of bacteria (relative to molecules in a solution) cause light to be refracted in a phenomenon known as scattering. This results in a deviation from Beer’s Law (unless the bacterial suspension is dilute) In bacterial suspensions less light is transmitted to the detector and the increased absorbance value is proportional to the cell density. (This is only valid in dilute suspensions where o.d. < 1) 2. Other deviations in Beer’s Law are observed in complex solutions and suspensions. 3. In mixtures, the absorption spectrum is a linear sum of the component spectra. Therefore: A = (ε*c 1 + ε*c 2 ) d Problem Solving As the head of a sunscreen company you wish to test the effectiveness of a new sunscreen (product 2) versus a competitive brand(product 1) which has an SPF of 20. The absorbance of Product 1 is double that of Product 2 when the thickness of Product 2 is 4 times that of Product 1. What is the SPF of your new sunscreen? Answer Sunscreen 2 has: 0.5 Absorbance, 4 times thickness Since A=εcd, the normal Abs of (sunscreen 2) is 0.5/4 = 0.125 of (sunscreen 1) T1 = 1/SPF = 1/20 = 0.05 A1 = -log(T1) = 1.3 A2=(0.125) (1.3) = 0.1625 T2 = 10-A2 = 10-0.1625 = 0.688 SPF2 = 1/T2 = 1/0.688 = 1.45 Enzyme Kinetics Enzymes are catalysts that accelerate the rate of chemical reactions They function by lowering the activation energy, but do not alter the overall energy of the reaction (∆ G) or the position of equilibrium Examples: pepsin, urease Reaction Rates The rate of an enzyme catalyzed reaction can be measured as a function of product formation (∆P/∆t) Alternatively, enzymatic rate can be measured as a function of substrate loss (∆S/∆t) The rate of product formation is usually preferred (much easier to quantify) Enzyme kinetic experiments measure the initial velocities or rates of reaction (linear portion of the curve) Graph of V vs. [S] The plot to the left shows the Vo Vmax= maximum rate of activity Km= concentration of substrate at half maximal activity Vmax/Km = physiological efficiency (initial rate) values at varying substrate concentrations for a given enzymatic reaction. Vmax represents the maximum rate of activity and occurs when the enzyme is saturated with substrate. Km is the [S] at half maximal velocity and represents the binding affinity of the enzyme At low [S], V0 is nearly proportional to [S] At high [S], V0 approaches Vmax and becomes independent of substrate concentration Reaction Rate Equations Reaction scheme for an enzyme-catalyzed reaction: The rate of ES formation can be represented as follows: Two assumptions can be made to further simplify this equation: The initial concentration of product ([P]) is ≈ 0. Therefore the term k-2 [E][P] can be eliminated. 2. The rate of change of [ES] is ≈ 0 (pseudo steady state) 1. Steady State Assumption The Steady State assumption states that the concentration of ES complex remains constant over time Thus, the rate of formation of ES is equal to the rate of degradation and ∆[ES]/∆t = 0 Simplified Reaction Mechanism Simplified Rate Equation Michaelis Constant From steady state, we have: K1[E][S] = K-1[ES] + Kcat[ES] (Formation = Destruction) Isolate for [ES]: K1[E][S] = (K-1 + Kcat)[ES] Rearrange: [E][S]/[ES] = (K-1 + Kcat)/K1 = KM Deriving the Michaelis-Menten Equation Start with the simplified rate equation Recall : Substitute [E] = [Et]- [ES] and rearrange the equation as follows: Recall: Final Form! Starting Equations [ET] = [ES] + [E] K1[E][S] = K-1[ES] + Kcat[ES] KM = (K-1 + Kcat)/K1 Vo = kcat[ES], Vmax = kcat[ET] Total Enzyme Steady State Michaelis Constant Reaction Rate Get rid of all the E’s and ES’s! Start by solving Steady State for [E] and plugging into the Total Enzyme equation. Derive it Yourself! Michaelis-Menten Equation *Learn the derivation! Km: an "aggregate" constant (sum of rate constants for breakdown of ES divided by rate constant for formation of ES): The Michaelis-Menten equation explains the Vo vs. [S] curve: 1. At very low [S] ([S] << Km), Vo approaches (Vmax/Km)[S]. Vmax and Km are constants, so linear relationship exits between Vo and [S]at low [S]. 2. 3. When [S] = Km, Vo = 1/2 Vmax At very high [S], ([S] >> Km), Vo approaches Vmax (velocity independent of [S]) Plotting the Data Lineweaver-Burke Plot Slope = Km/Vmax Yint = 1/Vmax Xint = -1/Km Eadie-Hofstee Plot Slope = -Km Yint = Vmax Xint = Vmax/Km ** The problem with the Lineweave- Burke plot is that the data points are not well distributed. The Eadie-Hofstee plot provides a more even weighting of the data points. Turnover Rate and Efficiency The turnover rate of an enzyme, k2, is defined as the maximum number of molecules of substrate that an enzyme can convert to product per unit of time. k2= Vmax/[E]T The turnover rate is measured in units of sec-1 or min-1 The physiological efficiency of a reaction is defined as Vmax/ Km Sample Calculation Practice Questions 1. If you double enzyme concentration, how is the Vmax value affected? How is the Km value affected? 2. The Vmax of an enzymatic reaction is 20mM per min and you have a total enzyme concentration of 0.1 M. What is the turnover rate of the enzyme? 3. The activity of 6 ug of enzyme is measured by absorbance in a 1ml, 1cm cuvette. The change in absorbance is measured as 0.18 O.D./min and the molar extinction coefficient is measured at 220 M-1 cm-1 . Calculate the activity in umol/min/ug. 1. Vmax = kcat[ET], Doubles. Km = (k-1 + kcat)/(k1), no change 2. Vmax = kcat[ET] Kcat = Vmax/[ET] = (20 mM/min)/0.1 M = 0.2 min-1 = 12 s-1 3. OD/min µmol/min A=εcd c = A/(εd) = 0.18/(222M-1cm-1 x 1 cm) = 810.8µM (810.8µM/min)/(6µg) = 135µM/min/µg Enzyme Inhibition Inhibitors are compounds that block enzyme activity Two main types of inhibitors are competitive and non-competitive. Competitive Inhibitors: Compete with the substrate for the enzyme active site These inhibitors function by decreasing the binding affinity of an enzyme for its substrate Km increases; Vmax remains unchanged Non-competitive inhibitors : Inhibitor binds to a site other than the active site and does not compete with the substrate for binding These inhibitors function by decreasing the turnover rate of an enzyme Vmax decreases; Km remains unchanged Competitive vs. Non-Competitive Inhibition Competitive Inhibition: Non-competitive Inhibition: Reaction Scheme Reaction Scheme **Enzyme binds either I or S Lineweaver-Burke Plot **Enzyme can bind both I and S Lineweaver-Burke Plot Inhibition Table Type of Inhibition Km V max Physiological efficiency None Km Vmax Vmax/Km Competitive Kmapp = Km (1 + [I]/Ki ) Vmaxapp = Vmax (Vmax is unchanged) Vmax/ Km*(1 + [I]/Ki ) Non-competitive Kmapp = Km (Km is unchanged) Vmaxapp = Vmax/(1 + [I]/Ki ) Vmax/ Km *(1 + [I]/Ki ) *Note: Ki is the concentration at which the inhibitor is 50% effective (1 + [I]/Ki ) = α (inhibition factor) Tips for enzyme problems! Use your units to help avoid errors Moles are not Molar (mol/L) Be explicit with conversion factors E.g. 1 = 1 ml/ 10-3 L Check the units in your answer Do they make sense? Are the values too high or too low? Use reasonable precision Test your understanding 1. Why would lowering enzyme concentration be equivalent to adding a non-competitive inhibitor? 2. The Vmax of a reaction is 33.2 mol/L/year. What is the Vmax in umol/ml/sec? 3. In the presence of an inhibitor ([I] = 1 mM), the Km is increased by a factor of 3. Calculate the Ki value. What type of inhibition is this? Challenge Question The kinetics of an enzyme are measured as a function of substrate concentration in the presence and in the absence of 2 mM inhibitor (I). Plot the data as an Eadie-Hofstee plot (a) What are the values of Vmax and KM in the absence of inhibitor? In its presence? (b) What type of inhibition is it? (c) What is the binding constant of this inhibitor? (d) If [S] = 10 μM and [I] = 2 mM, what fraction of the enzyme molecules have a bound substrate? A bound inhibitor? Hint: fractional [ES] = ratio of occupied sites to active sites= V0/ Vmax Solution (a) In the absence of inhibitor, Vmax is 47.6 μmol minute-1, and KM is 1.1 × 10-5 M. In the presence of inhibitor, Vmax is the same, and the apparent KM is 3.1 × 10-5 M. (b) Competitive. (c) 1.1 × 10-3 M. (d) fES is 0.243, and fEI is 0.488.