Chemistry Graduate Research - Central Washington University
advertisement

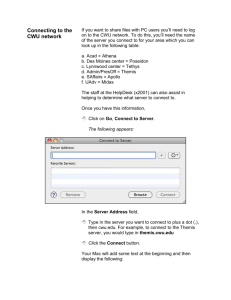
Research Programs A completed research project is the cornerstone of the graduate program in chemistry. Following is a list of graduate faculty mentors and their respective areas of interest from which a student can choose: Gil Belofsky (belofskyg@cwu.edu) Isolation and structural characterization of organic compounds from plants. Medicinal and pharmacological applications of natural products. Department of Chemistry Central Wa. University MS options: • • • Thesis Non-thesis project BS/MS 5-year program Timothy Beng (timothyb@cwu.edu) Development of synthetic methodology for efficient and expedient construction and functionalization of nitrogen and oxygencontaining heterocycles, for eventual application in natural and unnatural product synthesis. Anthony Diaz (diaza@cwu.edu) Luminescence, energy transfer and degradation processes in solid state luminescent systems. Levente Fabry-Asztalos (fabryl@cwu.edu) Design and synthesis of inhibitors against therapeutically important enzymes. Yingbin Ge (yingbin@cwu.edu) Theoretical chemistry and computational studies of the properties of nanoclusters, nanocatalysis, astrochemical species and reactions. Last revision in Feb, 2016 Anne Johansen (johansea@cwu.edu) Chemical composition of ambient aerosol particles and surface waters in the context of global climate and public health. Analysis of wines. Todd Kroll (kroll@cwu.edu) Identification and characterization of proteinprotein interactions regulating the size, position and lamination of functional areas of mammalian neocortex. Martha Kurtz (kurtzm@cwu.edu) Chemistry education, environmental-based integrated inquiry, critical thinking skill assessment. JoAnn Peters (petersj@cwu.edu) Laboratory and computational studies of organic reactive intermediates. Robert Rittenhouse (rittenhr@cwu.edu) Computational studies of the structure and dynamics pertaining to substrate binding in DNA repair enzymes. Dion Rivera (riverad@cwu.edu) Spectroscopic investigations of macromolecular complexes of polyelectrolytes and surfactants in solution and at nanoparticle interfaces. Tim Sorey (soreyt@cwu.edu) Development and integration of instrumental measurement technology in educational laboratories and the synthesis of educational strategies that support their use. Carin Thomas (thomasc@cwu.edu) We study the effects of environmental contaminants and nanoparticles on biological systems, examining how these chemicals interact with reactive oxygen species and antioxidants to affect cellular and mitochondrial function. Belofsky Research Why Natural Products? Track record of success Discovery process High-demand skills Field work component Rationale for Selection of Organisms of Interest Ecological: suspected chemical defenses or communication Geographic: “extreme” or difficult to access environments Some Current Projects: (Other Dalea species are also currently under investigation) Collaborators: Blaise Dondji and students (CWU, Biology). Dalea ornata Status: Manuscript in preparation; ten compounds reported. Activity: Anti-hookworm (anthelmintic). Dalea emoryi (Psorothamnus emoryi) Status: Compound isolation and characterization is ongoing… Activity: A compound that is quite active against hookworm has been revealed, the known compound dalrubone: Taxonomic: infrequently or never before studied Ethnographic: traditional or folklore uses We have been heavily focused on the chemistry of the plant genus Dalea, after investigating many different genera using the above rationale. We also discovered a new compound we named emoryan that is weakly active: chromatography column showing the purification of dalrubone Diaz Research Group – electron migration and trapping in luminescent materials Dr. Diaz’s research involves the study of electronhole (e-h) pair transport and trapping in doped luminescent materials under vacuum ultraviolet (VUV) excitation. Excitation by VUV radiation leads to the formation of an e-h pair in the host. In order for luminescence to occur this e-h pair must be trapped by the rare earth dopant. However, the electron may also be trapped by bulk killers (impurities or defects), or it may be lost to surface states. In this figure YBO3 is the host and Eu3+ is the dopant. The purpose of our research is to quantify the fate of the e-h pair after absorption of a VUV photon takes place. Above is another view of the process, which shows the electronic states involved. Once created, the e-h pair migrates through the lattice until it is trapped by killers or by a dopant. Dopant states are in blue, and loss to killers is indicated by the dashed line. The overall efficiency of host excitation once a photon is absorbed is given by hhost = ht*hqe, where ht is the transfer efficiency and hqe is the quantum efficiency of the dopant after the e-h pair is trapped. The transfer efficiency is then hhost/hqe. These quantities are determined spectroscopically via absorbance and excitation measurements – essentially comparing the amount of light the material absorbs to the amount of light emitted by the dopant after absorption. Diaz Research Group – electron migration and trapping in luminescent materials Once transfer efficiency data are collected they are modeled using the equation on the left. The transfer efficiency is simply the ratio of the rate of transfer to dopants (also called “activators”) divided by the combined rate of trapping by killers and activators. The multiplier Sloss is equal to 1 when no energy is lost to the surface, and approaches zero as more surface loss takes place. If transfer efficiency data are collected for a series of dopant concentrations, the a/b ratio and the value of Sloss can be determined. Theoretical curves are shown below on the left, while recent data on nanocrystalline YBO3:Eu3+ are shown on the right. With particle sizes > 500 nm no surface loss is observed, while at 25 nm more than 40% of absorbed energy is lost to the surface. Fabry Research Group Design and Synthesis of Novel Enzyme Inhibitors My research group is interested in addressing biologically and medically important questions. The focal point of our research is the design and synthesis of small molecule inhibitor scaffolds against therapeutically important enzymes. Our goal is to find orally active inhibitors that could become lead compounds for further drug discovery. During this process, we are developing new and improving already known synthetic chemistry methodologies. To achieve our goals we use all the modern tools of medicinal chemistry and organic synthesis. Medicinal Chemistry Organic Synthesis Computer Modeling HO B H N N Ac-Ser-Leu-Asn-HN OH O OH B R1 N R2 Pharmacology Dr. Levente Fabry-Asztalos; fabryl@cwu.edu; (509) 963-2887; SCI 302F O Fabry Research Group Design and Synthesis of Novel Enzyme Inhibitors Also, as a joint research effort with a computer science group we develop and extensively test new molecular modeling and computational chemistry techniques. This endeavor centers on molecular modeling, as well as computational intelligence techniques, which include neural networks, fuzzy systems, evolutionary computation, and biology inspired computational models. 2.5 log(Predicted_IC50) 2 S 1.5 1 N O H N N N H 0.5 R 2 = 0.7673 O 0 -1 -0.5 0 0.5 1 1.5 2 N H N O S O 2.5 -0.5 -1 log(Actual_IC50) Pharmacology 25 Distance (Å) 20 15 10 5 0 AR G LE 8 U AS 23 P2 AS 5 P VA 29 L3 IL 2 E PR 47 O VA 81 L8 AR 2 G LE 8 U2 AL 3 A2 AS 8 P VA 29 L3 IL 2 E PR 50 O 8 IL 1 E8 4 -1.5 OH 1D4Y to 1HPV 1D4Y to 1HXB 1D4Y to 1HXW 1D4Y to 1MUI 1D4Y to 1OHR 1D4Y to 2BPX 1HPV to 1HXB 1HPV to 1HXW 1HPV to 1MUI 1HPV to 1OHR 1HPV to 2BPX 1HXB to 1HXW 1HXB to 1MUI 1HXB to 1OHR 1HXB to 2BPX 1HXW to 1MUI 1HXW to 1OHR 1HXW to 2BPX 1MUI to 1OHR 1MUI to 2BPX 1OHR to 2BPX Computer Modeling Atoms Medicinal Chemistry Organic synthesis Dr. Levente Fabry-Asztalos; fabryl@cwu.edu; (509) 963-2887; SCI 302F Chemistry with Computers Astrochemistry in Ice Yingbin Ge From water to water oxide to hydrogen peroxide Europa Callisto Ganymede Ptn C3H8 + 1/2O2 C3H6 + H2O Bulk silicon Si nanoclusters emit bright light 7 My recent presentations and research interests are posted on http://www.cwu.edu/~yingbin/research.html My CV including a publication list is posted on http://www.cwu.edu/~yingbin/cv/cv.pdf My questions to you are which one of my papers or projects interests you the most and why? Johansen Research - Current Projects 1. Iron in Aerosol Particles (NSF) – Implications on Global Climate and Human Health • • Crustal/Marine Anthropogenic 2. Pollution Monitoring at Mt. Rainier and North Cascades National Parks (NPS) • Precipitation • High elevation lakes 3. Chemistry of Faulty Wines • Analyses • Method development (Continuing Ed. And Biology) Nature of the Work - Examples Field Aerosol Collector Collect particles in 4 size fractions at sea and regionally. Laboratory Solar Simulator Study photochemistry in synthesized and ambient aerosols. QUANTITATIVE ANALYSIS Instruments in Chemistry, Geology, EMSL IC, Chemiluminescence FIA, ICPMS, XPS ExamplesFactors of new Kroll Lab: Graded Expression of Transcription Methods and results flavanoids found… Regulates Neocortical Arealization Graded Expression: A gene being turned on in a high to low gradient. Transcription Factors: Class of proteins that regulate the turning on and off of specific genes Neocortical Arealization: The process of dividing the neocortex into functional units The neocortex of all mammalian species Structure determination of unknown compounds have four primary areas, the Motor (M), Extensive 1D and 2D NMR spectroscopy Somatosensory (S1), (V1), and Recent upgrade of our ownVisual 400 MHz instrument to runAuditory advanced(A1) 2D experiments like HSQC & HMBC High resolution mass spectrometry The sizes of these areas are different in Sent to the University of Iowa different individuals………Why? We continue to find new, interesting and active compounds from this plant genus! Kroll Lab: Graded Expression of Transcription Factors Regulates Neocortical Arealization Altering the concentration gradients of any of these transcription factors results in predictable changes in the size of neocortical areas: normal Emx2 reduced Emx2 change in gradient change in area sizes but, there are always clear boundaries separating the areas The big question now are: 1) How are these boundaries established 2) How do these transcription factors transmit positional information within the cells We are attempting to answer these questions by finding the proteins to which these transcription factors interact. Chemical Education Dr. Martha J. Kurtz Current Research Interests: Critical Thinking Skills Assessment through Community-based Inquiry Secondary Science Classroom Practice related to Elements of Effective Science Instruction Effectiveness of STEP Program in recruiting and retaining STEM majors Environment-based Integrated Learning in K-12 Schools Misconceptions in Science Chemical Education Research Knowledge and skills developed through education research Human subjects protocols Qualitative and quantitative research methods Controlling variables in studies of human behavior Applied statistics Understanding of effectiveness of various teaching strategies Applications of Computational Chemistry to the Mysteries of DNA Repair Rittenhouse research rittenhr@cwu.edu substrate primer DNA Mg2+ ions Solvated enzyme/gapped-DNA/ddCTP complex studied via molecular dynamics simulation Rittenhouse research rittenhr@cwu.edu MD simulation = all atoms + forces + thermal energy + time Rivera Research Group Investigation of macromolecular complexes and their interactions with guest molecules. H2O Polyelectrolyte/surfactant Complex (PSC) Goals: To understand how the PSC interacts with guest molecules. Understand the effects of the structure of the guest molecule on the its interaction with the PSC. Understand the influence of different surfactants on the formation of the PSC. TiO2 (s) Analytical Techniques Used: ATR-FTIR, UV-vis, quartz crystal microbalance, and surface tension measurement. Since macromolecular systems are inherently complex multivariate data analysis techniques such need to be applied to the acquired data in order to fully understand the systems. Example of a constraint applied to the UV-vis data set. - Matrix of Dye Spectra Original Data Matrix = Matrix with Dye removed 2 0.35 1.8 2 0.3 1.6 0.25 1.5 1 0.2 = 0.15 Constrained Absorbance 1.4 1.2 1 0.8 0.6 0.1 0.5 0.4 0.05 0 200 250 300 350 400 450 500 550 0 0.2 0 200 250 300 350 400 450 500 550 600 200 250 300 350 wavelength (nm) 400 450 500 Sorey Research Group – Timothy L. Sorey, PhD. Central Washington University soreyt@cwu.edu (continued) Research student example of development and test of computer-based instrument 1) Schematics of comparison polarimeter 2) Build and test of instrument for change in phase (Φ) between the standard and sample polarimeter cells Top View Legend to Apparatus: (a) (a) Removable LED light source (b) Sample Cell (c) Standard Cell (d) Stationary Polarizer Film (e) Rotating Polarizer Film (f) Phototransitor (g) Stepper Motor (h) Data Acquisition (i) Computer (f) (b) (g) (a) (c) Note: Dashed lines (d) (f) (e) (h) Power Supply (i) Side View End View (e) (d) (c) (End View) (b) (g) (c) (g) (a) (f) (e) (h) Power Supply (i) This is the “End View” that is drawn from a perspective behind the stepper motor, (g), towards the Rotating Polarizer Film, (e). 0.00g/10mL D-Fructose 2.00g/100mL D-Fructose 3) Calibration and validation of polarimeter with D-Fructose Concentration (g/10mL) -0.0211 0.00 0.117 0.50 0.285 1.00 0.367 1.50 0.555 2.00 0.5 Delta Phi Delta Phi (ΔΦ) Delta Phi versus Concentration - D-Fructose 0.6 y = 0.2808x - 0.0202 R² = 0.9903 0.4 0.3 0.2 0.1 0 -0.1 -0.1 0.4 0.9 Concentration (g/10mL) 1.4 1.9 Thomas Research Group: Effects of Environmental Factors on Mitochondrial Function and Reactive Oxygen Species Generation 2 1 2 HO 2 µM 1.5 0.5 0 -0.5 -500 0 500 1000 Time (s) 1 e- • Cellular respiration and ATP synthesis • Reactive Oxygen Species (ROS) • Antioxidant and Repair processes • Cell Signaling • Apoptosis: Cell Death O2· − H2O2 Fe2+ 4 e· OH 1500 2000 Mitochondrial Energetics & ROS Aerobic organisms have engineered antioxidant defenses against ROS Superoxide Dismutase (MnSOD) 2O2.- + 2H+ H2O2 + O2 Glutathione Peroxidase (GPx) GSH = intracellular thiol H2O2 + 2 GSH 2 H2O + GSSG Glutathione Reductase NADPH, H+ + GSSG 2 GSH + NADP+ Nicotinamide Nucleotide Transhydrogenase NADH, H+ → NAD+ + NADPH, H+ facilitates GSH recycling and removal of H2O2 If you are interested in the research of our graduate faculty members, please contact them via email or phone to ask for more information or make an appointment. Their contact information can be found at http://www.cwu.edu/chemistry/faculty 24