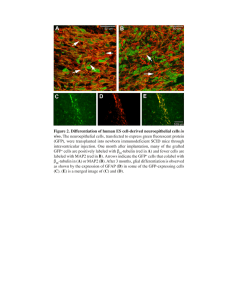
GFP protein purification lab
advertisement

GFP protein purification lab What are we trying to do • 7.3– Use E.coli that contains pGLO plasmid w/ GFP gene to express GFP protein 2ml of culture What are we trying to do • 7.3– Use Lysozyme enzyme break open bacterial cells to release GFP protein (TTh) – Spin down cellular debris, GFP protein now in supernatant. Move GFP containing supernatant to new MCT. Add high salt concentration binding buffer (decreases solubility of hydrophobic proteins like GFP (MWF) What are we trying to do • 7.3– Use Hydrophobic interaction column to elute proteins based on hydrophobicity • GFP is a relatively Hydrophobic protein based on its amino acids. Therefor it holds readily to the hydrophobic resin beds in a high salt concentration, but when the salt concentration is reduced the GFP goes from adhering to the beads to being in solution…and dripping out of the HIC column What are we trying to do • 7.3- extension … using SDS- page gel to analyze how purified the GFP protein became by using HIC column. – Protein Standards…by Kilodalton…why not by number of amino acids? – If we want to determine how purified the GFP was after 7.3, what do we also need to run in the gel? • hint: we did not make a lysate of untransformed E.coli Making a semi log curve mass Distance Basics of procedure • 1protein:1 Laemmli buffer – Compensates for R group variability…Laemmli buffer makes makes whole protein negatively charged • How? SDS (sodium dodecil sulfate) makes protein negative Basics of procedure • 1protein:1 Laemmli buffer – Compensates for R group variability…Laemmli buffer makes makes whole protein negatively charged • How? SDS (sodium dodecil sulfate) makes protein negative – Run 10 microliters in SDS-page gel in a vertical Electrophoresis system w/ 1x TGS (tris Glycine SDS*) *soapy • 200V • 30 min – Stain 1hr Biosafe Coomassie (protein Gel stain) – Destain overnight in dwater- on rocker Mini-Protean Tetra cell