Other structure programs
advertisement

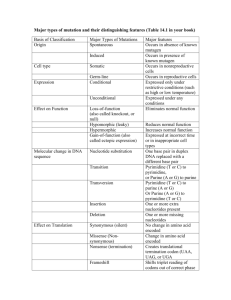
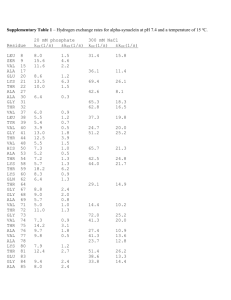
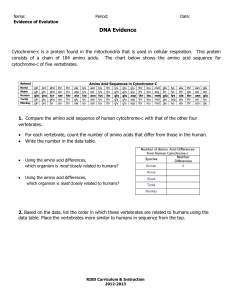
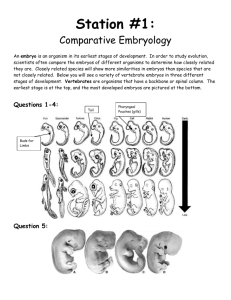
Other structure programs Insight II Swiss PDB-Viewer QUANTA/CHARMm and Quanta 2005 for proteins SYBYL with FUGUE and FlexX for drugs O or FRODO for x-ray MolMol for NMR PovChem, Rasmol, CHARMm Ranges Zones give the residue range: 17 or 19 20 0 but not 17 19 20 0 only P than you can pick a range in the graphic window Ranges PROT WATER DRUG All proteins All waters All drugs, ligands, ions NUC All nucleic acids SUG All sugars ALL All residues HELIX All helical residues STRAND All strand residues (-SHEET takes same ranges) ION All ions OXT All C-terminal oxygens <RESNAM> All residues of a certain type Start with – (minus sign) to remove things from selection. I.e. TOT –SUG gives everything except sugars. Numbering You can number directly a residue for example 27 in 3ERA the LYS (or with O27 if you need the number in brackets) but if LYS is not in m1 but in m3 you can number it as S2 27 or as U2 27. Try the numbering with LISTA WHAT IF> getmol 1boy Do you mean: /g/data9/data/pdb/1boy.brk Y/N ? y Give the set-name (1boy.brk) : 1 10 THR THR 11 20 THR TRP 21 30 LEU GLU 31 40 VAL TYR 41 50 GLY ASP 51 60 THR ASP 61 70 ILE VAL 71 80 ALA ARG 81 90 VAL GLU 91 - 100 LEU TYR 101 - 110 TYR LEU ..................................... WHAT IF> %shosou ASN LYS TRP THR TRP THR LYS VAL SER GLU GLU THR SER GLU VAL LYS GLU ASP PHE THR ASN THR VAL THR PRO GLN SER CYS VAL SER GLY SER ASN ALA ASN LYS ILE LYS ASP LYS TYR SER PRO LEU ALA PHE PRO SER CYS LEU GLN PRO ALA GLU GLY TYR LYS VAL THR PHE THR THR ALA GLY PHE GLN ASN THR ASN LYS TYR ASP TYR GLY GLU THR PRO LEU ILE GLN SER THR GLU LEU ASN PRO PRO THR Content of the SOUP. See the writeup for an explanation. Molecule 1 2 3 Range 1 ( 3) 212 ( 213) 213 ( HOH ) WHAT IF> Type Set name 211 ( 213) Protein 212 ( 213) E O2 <211 213 ( HOH ) water ( 110) SET_1boy.brk SET_1boy.brk SET_1boy.brk Three letter codes for menue The following three letter codes are associated with entire menus. ACC Has to do with ACCessibility BFT B-FacTor BLD BuiLD (mainly protein) CHK CHecK CHI Torsion angle CLU CLUster of 3D related residues COL COLour DG Distance Geometry rotamer and loop search DIG Digitalization (reconstruction from stereo plots) FAM FAMily, range of covalently connected residues GRA GRAphics GRI GRIn and GRId GRO GROmos H2O Water HBO Hydrogen BOnd (potential hydrogen bonds) HB2 Hydrogen Bond 2nd version (optimal hydrogen bonds) HSP HSsP (multiple sequence alignment files) HST Helix, Strand, Turn (in other words: secondary structure) LAB LABel (not picked label, but label in MOL-item) MAP 3D electron density, property or probability distribution MAP NEU NEUral net NMR Nuclear Magnetic Resonanced Three-letter codes for options 3 letters: DEL, INI, ON/OFF, TAB,ROW GRA,SHO but DG pairs of options: GET/MAK, SAV/RES exceptions: SIMPLE DIRECT Colours The correspondence between numbers and colours is: 1 Blue 30 Blue-ish purple 60 Purple 90 Red-ish purple 120 Red 150 Orange 180 Yellow 190 Light brown 220 Soft green 240 Green 270 Funny green 300 White-ish green 330 Light blue 360 Blue Also you can type: RED, GREEN, YELLOW, BLUE,PURPLE, ORANGE, CYAN, MAGENTA. LAST COLOUR COUNTS