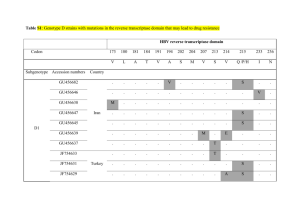
HIV Drug Resistance Training
advertisement

HIV Drug Resistance Training Module 6: Sequencing Interpretation and Analysis 1 Topics Understanding Mutations Identifying Resistant Mutations Phylogenetic Analysis Subtype Determination 2 Objectives Identify information needed by physicians and policy makers to make treatment decisions on a population basis. Describe algorithms used to translate sequence results to drug susceptibility predictions. Analyze results to determine HIV subtypes. Identify how to complete phylogenetic analysis. 3 understanding mutations What are the different types of mutations? How are mutations identified? 4 Genotype Interpretation: The Challenge K65R K70R E44D L74V M41L V118I V75M T215Y K219N Q151M T215F M184V K103N T69SSA F227L K219E K101P G190A V106M “Sensitive” to ?? “Resistant” to ?? “Maybe” to ?? I50V M46I G48V A71V V32I V82A I54V I47V L90M D30N V82F N88S I84V 5 Mutation Molecular definition: change in nucleic acid sequence compared to a reference sequence Biological definition: change in nucleic acid sequence that results in a change in structure or function of the nucleic acid or a resulting protein Codon AAA GAC AGT AAA AAC AGT Silent Mutation AAA GAC AGT AAA AAC AGC Lys Asp Ser (K) (D) (S) Lys Asn Ser (K) (N) (S) 6 Mutational Nomenclature G48V Wild-type (wt) amino acid (consensus or reference) Mutant amino acid Codon position PR: 1-99 amino acids RT: 1-540 amino acids L10L/I (mix of wt and mutant) V82A/F (mix of 2 mutants) 7 Effect of Nucleotide Changes Nucleotide changes (mutations) Changes in amino acid sequence of a protein Changes in structure/function of the protein (e.g. PR or RT) Changes in ability of drug to inhibit target enzyme (resistance) 8 Types of Mutations Polymorphisms – Naturally occurring mutations, not selected by drugs (but can influence susceptibility) “Primary” mutations – Directly affect drug binding, present near active site – Appear first in pathway to resistance – Not present in virus not exposed to drug pressure “Secondary” mutations – Compensate for fitness defects – Do not usually confer resistance on their own but modulate susceptibility – May include polymorphisms that are found more frequently in resistant viruses 9 Residues Associated with HIV PI Resistance Primary 48, 50, 82, 84… Flap 46, 47, 53, 54… Secondary 10, 20, 32, 36, 63, 71… How are Drug Resistance Mutations Identified? Researchers identify mutations selected by the drug during in vitro selection, phase I studies, site-directed mutagenesis BUT… Drug resistance mutations identified during drug development (esp. in vitro) may not be the most relevant mutations in clinical settings. Mutations that are sufficient to cause drug resistance may not be necessary to effect drug resistance. There may be cross-resistance due to mutations selected by related drugs. 11 Discussion What are the different types of mutations? How are mutations identified? 12 identifying resistance mutations How can you identify which mutations are responsible for drug resistance in your population? 13 Basis for Drug Resistance Knowledge Genotypic-phenotypic correlations on laboratory isolates (often confirmed by site-directed mutagenesis studies) 2. Genotypic-phenotypic correlations on clinical HIV-1 isolates 3. Correlations between HIV-1 genotype and the treatment history of patients from whom sequenced virus isolates are obtained 4. Correlations between HIV-1 genotype and the virologic response to a new treatment regimen. 1. 14 http://hivdb.stanford.edu/pages/documentPage/drm.html Selected Resistance and Cross-Resistance Exposure to new drug X Mutations A, B, C, D, E, F Exposure to other drugs from same class Mutations A, D, E, G, H, I Exposure to drugs from other classes Mutations J, K, L… Some mutations contribute to resistance (decrease susceptibility) Others may suppress resistance (increase susceptibility) 15 Sources of Genotype Interpretation Information IAS-USA (Hirsch et al., JAMA 2000; annual+ updates) – Expert consensus; updated frequently Stanford (R. Shafer), www.HIVResistance.com – Comprehensive, updated frequently, good notes Resistance Collaborative Group (DeGruttola et al., 2000) – Initially used in GeneSeq assay, not currently used GART (Baxter et al., 2000), VIRADAPT (Durant et al., 1999) – used in prospective clinical trials TruGene, ViroSeq, other test providers ABL/Virology Networks European Resistance guidelines ANRS, other EU country-specific guidelines 16 International AIDS SocietyUSA* Drug Resistance Mutations Group IAS-USA information available at: http://www.iasusa.org/resistance_mutations/index.html. *The International AIDS Society–USA (IAS–USA) is a not-for-profit, HIV clinical specialisteducation organization. It is entirely different from and not affiliated with the International AIDS 17 Society (Stockholm, Sweden). Multi-NRTI Resistance Mutations (IAS-USA) 18 Johnson et al. Topics HIV Med. 16(5): 138, December 2008. Updated on www.iasusa.org. © 2008. The International AIDS Society–USA Individual NRTI Resistance Mutations: IAS-USA 19 Johnson et al. Topics HIV Med. 16(5): 138, December 2008. Updated on www.iasusa.org. © 2008. The International AIDS Society–USA NNRTI Resistance Mutations (IAS-USA) 20 Johnson et al. Topics HIV Med. 16(5): 138, December 2008. Updated on www.iasusa.org. © 2008. The International AIDS Society–USA PI Resistance Mutations (IAS-USA) 21 Johnson et al. Topics HIV Med. 16(5): 138, December 2008. Updated on www.iasusa.org. © 2008. The International AIDS Society–USA NRTI Resistance Mutations (Stanford) Mutations in bold red are associated with higher levels of phenotypic resistance or clinical evidence for reduced virological response. 22 http://hivdb.stanford.edu/cgi-bin/NRTIResiNote.cgi NNRTI Resistance Mutations (Stanford) Mutations in bold red are associated with higher levels of phenotypic resistance or clinical evidence for reduced virological response. 23 http://hivdb.stanford.edu/cgi-bin/NNRTIResiNote.cgi PI Resistance Mutations (Stanford) Mutations in bold have been shown to reduce in vitro susceptibility or in vivo virological response. Mutations in bold underline are relative contraindications to the use of specific PIs. 24 http://hivdb.stanford.edu/cgi-bin/PIResiNote.cgi Mutations vs. Rules Mutations: amino acid changes in a patient sequence relative to some reference (e.g. HXB2, NL4-3, or consensus B) – Some mutations are associated with drug resistance Rules: specify certain mutations or combinations of specific mutations as being determinants of drug resistance – e.g. “T215Y plus 2 or more TAMs” Simple lists of mutations (e.g. IAS-USA, HIVResistanceWeb) are NOT rules! 25 Stanford HIVdb Interpretation (1 of 2) 26 Stanford HIVdb Interpretation (2 of 2) 27 Discussion How can you identify which mutations are responsible for drug resistance in your population? 28 phylogenetic analysis What is phylogenetic analysis, and why is it useful? 29 Phylogenetic analysis QA (detection of contamination) Identify epidemiological linkages Study mutation pathways 30 Phylogenetic Analysis Compare a test sequence to others – Tested in same batch or over recent period of time – Collected at the same site – Related sequences with possible index-source relationship Phylogenetic relationships can be described quantitatively and use statistical methods to test significance of linkages Challenging to decide on absolute thresholds to define 2 viruses as being "highly related" etc. (98%? 99%?) 31 Phylogenetic Tree Clusters of highly related sequences Reference sequences 32 100 7028 307l 100 100 C7018 5009 5043 5017 7008 T3046 E5025 3044 02_MP1211 84 02_DJ.DJ263 100 3031NV T3003 100 02_NG.IBNG 01_TH.CM240 98 01_TH.93TH253 01_CF.90CF402 84 A3_DDI579 A3_DDJ360 100 100 90 A3_DDJ369 A1_SE.SOSE7253 96 A1_UG.92UG037 A1_KE.Q2317 E5047 E5056 7085 100 7022 5011 100 G_BE.DRCBL G_SE.SE6165 G_FI.HH8793 06_95ML84 100 06_BFP90 06_97SE1078 100 K_97CD.EQTB11 K_96CM.MP535 F2_CM53657 100 F2_96CM.MP257 F2_96CM.MP255 100 05_X492 05_VI961 F1_BE.VI850 100 F1_FI.FIN9363 F1_BR.93BR020 78 D_UG.94UG114 D_99TCMN011 D_ZR.NDK 100 B_US.WEAU160 B_US.JRFL B_FR.HXB2R CRF02 CRF01 A3 A1 G CRF06 K F2 CRF05 F1 0.02 D B subtype determination What are subtypes and how are they determined? 34 HIV-1 Subtypes gag pol env 35 Global Distribution of HIV-1 Genetic Subtypes (2004) 36 Hemelaar et al. AIDS 2006, 20:W13–W23 Subtype Determination Simplistic Approach – Compare test sequence to set of reference sequences – Subtype of closest matching reference sequence is assigned to test sequence Reality: recombination between subtypes – PR matches one subtype; RT matches another – Circulating recombinant forms: found in unlinked individuals – Unique recombinant forms (URF) – Look for similarity to reference sequences using substrings of test sequence; "sliding window" 37 Circulating Recombinant Forms (CRF) CRF02_AG CRF06_cpx 38 http://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html Naturally Occurring Amino Acids in Non-B Subtype HIV-1 Position 98 179 181 190 10 20 36 46 69 71 93 NNRTI Amino Acid Subtypes G* C D*, I nd C Group O A* C PI I, V A, F, J, group O R, M, I Most non-B I, L Most non-B, group O I* C, F, AE K Most non-B V Group O L C * isolated observations nd = not defined 39 Adapted from Parkin and Schapiro, Antiviral Therapy 9: 3-12, 2004 Subtype-Specific PR Polymorphisms (Untreated Patients) 40 Kantor R et al. PLoS Medicine 2005 Apr;2:112 Subtype-Specific Treatment (PI)-Related Mutations 41 Kantor R et al. PLoS Medicine 2005 Apr;2:112 Are Subtype B Resistance Mutations also Treatment-related in Non-Bs? Each of the 55 known DR mutations occurred in at least one non-B isolate; 44 (80%) of these mutations were significantly associated with therapy in non-B isolates Most positions associated with DR in subtype B viruses are selected by antiretroviral therapy in one or more non-B subtypes as well. No evidence that non-B viruses develop mutations at positions that are not associated with resistance in subtype B viruses. Based on currently available data, global surveillance efforts and genotypic assessments of DR should focus primarily on the known subtype B 42 drug-resistance mutations. Kantor R et al. PLoS Medicine 2005 Apr;2:112 HIV-1 Subtype Influences Mutation Preference Drug NFV EFV d4T Subtype C L90M V106M K65R Subtype B D30N K103N TAMs Different subtypes may develop different profiles of mutations in response to the same treatment However, once detected, the interpretation of the meaning of these mutations is independent of subtype 43 Subtype Determination Subtype is not a critical factor for genotype interpretation Different prevalence of polymorphisms and secondary mutations Different mutational pathway preferences Geographic and/or epidemiological clustering 44 What about HIV-2? Naturally Occurring Amino Acids in SIV and HIV-2 Implicated in Modulation of Antiretroviral Drug Susceptibility (at Positions Selection form HIV-1 Knowledge Base) NRTI NNRTI PI FI (gp41) Position Amino acid Position Amino acid Position Amino acid Position Amino acid 69 N 101 A, E, P 10 V 42 Q 75 I 103 C, M, R 20 V 43 Q 118 I 106 I 32 I 210 N 179 T 36 I 215 S 181 I, V 46 I, V 219 E 188 F, L, W 47 V 190 A 71 I, V 227 Y 77 T 82 I Drugs impacted: NNRTI (all), PI (APV, ATV, TPV, DRV?) 45 Parkin and Schapiro, Antiviral Therapy 9: 3-12, 2004 Discussion What are subtypes and how are they determined? 46 Discussion What information do physicians and policy makers need to make treatment decisions on a population basis? How can we get them this information? What sources of information/interpretation would be useful to us? 47 Small Group Exercise Work in small groups. Read the directions Look through some sample sequences (five pages). Identify mixtures. 48 Example 1 49 Example 2 50 Example 3 51 Example 4 52 Example 5 53 Reflection What information do physicians and policy makers need to make treatment decisions on a population basis? How can we get them this information? What sources of information/interpretation would be useful to us? 54 Summary Understanding Mutations Identifying Resistant Mutations Subtype Determination Phylogenetic Analysis 55