Analysis of variance
advertisement
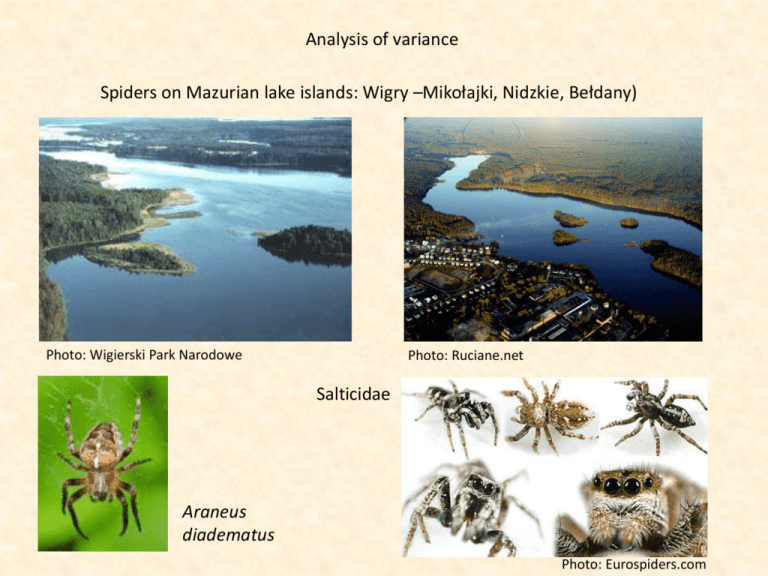
Analysis of variance Spiders on Mazurian lake islands: Wigry –Mikołajki, Nidzkie, Bełdany) Photo: Wigierski Park Narodowe Photo: Ruciane.net Salticidae Araneus diadematus Photo: Eurospiders.com Spider species richness on Mazurian lake islands Island Disturbance Species Górna E High 33 Kopanka High 34 Kopanka N High 32 Piaseczna High 38 Górna W High 29 Królewski Medium 51 Ostrów Wygryńska Medium 43 Maleńka Low 6 Ruciane - ląd Low 28 Mikołajki - ląd Low 75 Wierzba Low 47 Kamień Low 60 Mysia Wigry Low 49 Ordów Low 64 Koń Pristine 25 Mała Wierzba Pristine 27 Ośrodek Pristine 22 Śluza Pristine 19 Bryzgiel Pristine 21 Bryzgiel - ląd Pristine 46 Brzozowa L Pristine 31 Brzozowa P Pristine 30 Cimochowski Pristine 25 Grądzik C Cimochowski Pristine 31 Grądzik N Cimochowski Pristine 25 Grądzik S Krowa Pristine 34 Ostrów Pristine 93 Rośków Pristine 24 Walędziak Pristine 28 Wysoki Pristine 57 High 33 34 32 38 29 High Medium Low Medium 51 43 Low 6 28 75 47 60 49 64 Pristine 25 27 22 19 21 46 31 30 25 31 25 34 93 24 28 57 T-TEST Medium Low Pristine 0.145265 0.172254 0.931288 1 0.081749 0.211812 Bonferroni corrected High Medium Low If we use the same test several times with the same data we have to apply a Bonferroni correction. Single test p(nsig ) 1 p( sig ) n independent tests pExp (nsig ) (1 ptest ( sig )) n pExp ( sig ) 1 (1 ptest ( sig )) n 1 (1 nptest ( sig )) pExp ( sig ) nptest ( sig ) Exp 0.05 n Test T-TEST Medium Does species richness differ with respect to the degree of disturbance? Low Pristine 0.857544 0.862042 0.988548 1 0.846958 0.868635 Test 0.05 n Spider species richness on Mazurian lake islands Island Disturbance Species Górna E High 33 Kopanka High 34 Kopanka N High 32 Piaseczna High 38 Górna W High 29 Królewski Medium 51 Ostrów Wygryńska Medium 43 Maleńka Low 6 Ruciane - ląd Low 28 Mikołajki - ląd Low 75 Wierzba Low 47 Kamień Low 60 Mysia Wigry Low 49 Ordów Low 64 Koń Pristine 25 Mała Wierzba Pristine 27 Ośrodek Pristine 22 Śluza Pristine 19 Bryzgiel Pristine 21 Bryzgiel - ląd Pristine 46 Brzozowa L Pristine 31 Brzozowa P Pristine 30 Cimochowski Pristine 25 Grądzik C Cimochowski Pristine 31 Grądzik N Cimochowski Pristine 25 Grądzik S Krowa Pristine 34 Ostrów Pristine 93 Rośków Pristine 24 Walędziak Pristine 28 Wysoki Pristine 57 One way analysis of variance Sir Ronald Aylmer Fisher (1890-1962) sH2 x H sM 2 x M If there would be no difference between the sites the average within variance sWithin2 should equal the variance between the sites sBetween2 . s L2 x L sBetween2 sT2 sP2 x P F s 2Between 2 sWithin s 2Between sT2 s 2Between We test for significance using the F-test of Fisher with k-1 (Between) and n-k (Within) degrees of freedom. n-1 = n-k + k-1 df Total df Within df Between ni k sBetween 2 (x x i 1 i Total k 1 ) 2 k SS Between 2 s df Between W ithin i 1 SStotal SSbetween SSwithin (x j 1 i, j xi ) ni 1 n 2 SSW ithin dfW ithin sTotal 2 (x x i 1 i Total n 1 dftotal dfbetween df within )2 SSTotal dfTotal MS F t x1 x1 s12 s22 n1 n2 Welch test The Levene test compares the group variances using the F distribution. Variances shouldn’t differ too much (shouldn’t be heteroskedastic)!!! SS df MS Between MSW ithin The Tuckey test compares simultaneously the means of all combinations of groups. It’s a t-test corrected for multiple comparisons (similar to a Bonferroni correction) Observations 1 2 3 4 5 Group mean SSwithin Total SSwithin Total SSbetween Grand mean Grand SS Grand SS SSbetween+SSwithin F F-test A 0.08 0.71 0.19 0.51 0.73 0.445 0.131 0.070 0.065 0.004 0.082 4.11 13.96 1.08 1.00 0.14 0.79 0.32 0.12 18.07 18.07 18.14 2.118E-05 Treatments B C 0.19 0.83 1.21 0.71 1.97 1.10 0.19 0.11 0.19 0.30 0.750 0.611 0.319 0.046 0.216 0.010 1.484 0.244 0.314 0.250 0.312 0.096 SSbetween D 2.80 2.69 1.93 2.57 2.58 2.515 0.081 0.032 0.342 0.004 0.004 0.404 0.404 0.404 0.404 0.404 0.109 0.109 0.109 0.109 0.109 sBetween 2 (x x i i 1 k )2 k 1 (x j 1 i, j SS Between df Between SSW ithin dfW ithin xi ) 2 ni 1 i 1 2.96 2.61 0.72 2.23 2.24 Total ni 2 0.06 0.14 0.00 0.94 0.61 2.059 2.059 2.059 2.059 2.059 k sW ithin 0.80 0.02 0.79 0.79 0.79 0.220 0.220 0.220 0.220 0.220 n sTotal 2 (x x i 1 i Total n 1 )2 SSTotal dfTotal We include the effect of island complex (Wigry – Nidzkie, Bełdany, Mikołaiki) Island Complex Disturbance Species Górna E NBM High 33 Kopanka NBM High 34 Kopanka N NBM High 32 Piaseczna NBM High 38 Górna W NBM High 29 Królewski Ostrów NBM Medium 51 Wygryńska NBM Maleńka NBM Ruciane - ląd NBM Mikołajki - lądNBM Wierzba NBM Kamień Wigry Mysia Wigry Wigry Ordów Wigry Koń NBM Mała WierzbaNBM Ośrodek NBM Śluza NBM Bryzgiel Wigry Bryzgiel - lądWigry Brzozowa L Wigry Brzozowa P Wigry Medium Low Low Low Low Low Low Low Pristine Pristine Pristine Pristine Pristine Pristine Pristine Pristine 43 6 28 75 47 60 49 64 25 27 22 19 21 46 31 30 CimochowskiWigry Grądzik C Pristine There must be at least two data for each combination of groups. We use a simple two way ANOVA Island Complex Disturbance Species Maleńka NBM Low 6 Ruciane - ląd NBM Low 28 Mikołajki - lądNBM Low 75 Wierzba NBM Low 47 Kamień Wigry Low 60 Mysia Wigry Wigry Low 49 Ordów Wigry Low 64 Koń NBM Pristine 25 Mała WierzbaNBM Pristine 27 Ośrodek NBM Pristine 22 Śluza NBM Pristine 19 Bryzgiel Wigry Pristine 21 Bryzgiel - lądWigry Pristine 46 Brzozowa L Wigry Pristine 31 Brzozowa P Wigry Pristine 30 CimochowskiWigry Grądzik C Pristine 25 CimochowskiWigry Grądzik N Pristine 31 CimochowskiWigry Grądzik S Pristine 25 25 Krowa Ostrów Rośków Walędziak Wigry Wigry Wigry Wigry Pristine Pristine Pristine Pristine 34 93 24 28 CimochowskiWigry Grądzik N Pristine 31 Wysoki Węgieł Wigry Pristine 57 CimochowskiWigry Grądzik S Pristine 25 Krowa Ostrów Rośków Walędziak Wigry Wigry Wigry Wigry Pristine Pristine Pristine Pristine 34 93 24 28 Wysoki Węgieł Wigry Pristine 57 SStotal SS A SS B SS AxB SS error Main effects Secondary effects SS Complex SS Disturbance SS Complex Disturbance The significance levels have to be divided by the number of tests (Bonferroni correction) Spider species richness does not significantly depend on island complex and degree of disturbance. Correcting for covariates: Anaysis of covariance 100 y = 33.431x0.1917 R² = 0.7215 80 Species Island Complex Disturbance Area [ha] Species Górna E NBM 1 0.7 33 Koń NBM 4 0.5 25 Kopanka NBM 1 0.69 34 Królewski Ostrów NBM 2 6.15 51 Maleńka NBM 3 0.0003 6 Mała Wierzba NBM 4 0.4 27 Kopanka N NBM 1 0.18 32 Ośrodek NBM 4 0.09 22 Piaseczna NBM 1 0.63 38 Ruciane - ląd NBM 3 15 28 Mikołajki - ląd NBM 3 20 75 Śluza NBM 4 0.48 19 Górna W NBM 1 0.44 29 Wierzba NBM 3 0.78 47 Wygryńska NBM 2 0.67 43 Bryzgiel Wigry 4 0.2 21 Bryzgiel - ląd Wigry 4 16 46 Brzozowa L Wigry 4 3.81 31 Brzozowa P Wigry 4 2.32 30 Cimochowski Grądzik Wigry C 4 0.15 25 Cimochowski Grądzik Wigry N 4 0.14 31 Cimochowski Grądzik Wigry S 4 0.76 25 Kamień Wigry 3 3.13 60 Krowa Wigry 4 4.49 34 Mysia Wigry Wigry 3 1.55 49 Ordów Wigry 3 8.69 64 Ostrów Wigry 4 38.82 93 Rośków Wigry 4 0.56 24 Walędziak Wigry 4 0.76 28 Wysoki Węgieł Wigry 4 18 57 60 40 20 0 0 10 20 Area 30 40 Instead of using the raw data we use the residuals. These are the area corrected species numbers. The conmparison of within group residuals and between group residuals gives our F-statistic. 50 Disturbance does not significantly influence area corrected species richness Total residuals SStotal = SSbetween Within group residuals + SSerror We need four regression equations: one from all data points and three within groups. Before After SSbetween Medical treatment SSwithin n Repetitive designs In medical research we test patients before and after medical treatment to SStotal infer the influence of the therapy. We have to divide the total variance SSwithin (SStotal) in a part that contains the variance SSbetween between patients (SSbetween) and within the patient (SSwithin). SSError SStreat The latter can be divided in a part that comes from the treatment (SStreat) and the error (SSerror) k SS total ( xij x) 2 i 1 j 1 n SS between k ( Pi x) 2 i 1 n SStotal SSbetween SSwithin SSbetween SStreat SSerror df total df between df within df between df treat df error kn 1 n 1 n(k 1) n 1 k 1 (n 1)(k 1) k SS within ( xij Pi ) 2 k n (Tj x) 2 i 1 j 1 k SS treat n (T j x) 2 j 1 n k SS error ( xij Pi T j x) 2 i 1 j 1 F SStreat df error SSerror df treat j1 k n (x j1 i 1 ij Pi Tj x) 2 (n 1)(k 1) k 1 Before – after analysis in environmental protection Island Górna E Koń Kopanka Królewski Ostrów Maleńka Mała Wierzba Kopanka N Ośrodek Piaseczna Ruciane - ląd Mikołajki - ląd Śluza Górna W Wierzba Wygryńska Bryzgiel Bryzgiel - ląd Brzozowa L Brzozowa P Cimochowski Grądzik Cimochowski Grądzik Cimochowski Grądzik Kamień Krowa Mysia Wigry Ordów Ostrów Rośków Walędziak Wysoki Węgieł Mean P Grand Mean SStreat df Spring 26 19 21 50 6 25 28 16 34 22 43 12 19 29 26 15 44 22 29 C 19 N 29 S 14 37 19 32 37 77 21 14 32 27 23 1115.30 2 Summer 14 10 17 46 5 19 17 15 25 15 39 10 10 25 18 11 23 20 17 15 25 8 21 11 16 25 50 14 8 19 19 Autumn 22 16 15 47 4 21 23 12 29 13 26 7 11 23 26 14 28 13 23 17 29 14 37 13 29 25 57 17 13 19 21 Mean P 21 15 18 48 5 22 23 14 29 17 36 10 13 25 23 14 32 18 23 17 28 12 32 14 26 29 61 17 12 24 SS error df SStreat/ SSerror F p(f) SSError 15.2350 9.7935 11.4201 8.0776 29.2908 4.8073 4.5288 31.6569 0.6449 10.9042 120.5322 19.2222 5.0881 13.5529 18.3658 16.5768 98.0089 45.4914 6.8833 9.5781 17.3434 13.2847 93.9253 0.0904 60.1676 18.7889 193.4698 2.5835 9.1076 28.6667 917.0866 58 k SStreat n (T j x) 2 j 1 n k SS error ( xij Pi T j x) 2 i 1 j 1 dftreat = k-1 dfError = (n-1)(k-1) In the case of unequal variances between groups it is save to use the conservative ANOVA with (n-1) dferror and only one dfEffect in the final F-test. Mean P Grand Mean 1.2161338 SStreat 70.53576 2.953E-09 df 27 23 1115.30 1 19 21 SSerror 917.0866 df 29 SStreat/ 1.2161338 SSerror F 35.26788 p(f) 1.885E-06 Bivariate comparisons in environmental protection Species Residual Complex Area[ha] Species Predicted_Species Island 31.22156 1.778435 33 0.7 NBM Górna E 29.27129 -4.27129 25 0.5 NBM Koń 31.13556 2.864436 34 0.69 NBM Kopanka 47.35619 3.643813 51 6.15 NBM Królewski Ostrów 7.060143 -1.06014 6 0.0003 NBM Maleńka 28.04557 -1.04557 27 0.4 NBM Mała Wierzba 24.06496 7.935042 32 0.18 NBM Kopanka N 21.07064 0.929363 22 0.09 NBM Ośrodek 30.59729 7.402711 38 0.63 NBM Piaseczna 56.18315 -28.1831 28 15 NBM Ruciane - ląd 59.3686 15.6314 75 20 NBM Mikołajki - ląd 29.04312 -10.0431 19 0.48 NBM Śluza 28.5627 0.437301 29 0.44 NBM Górna W 100 31.87601 15.12399 47 0.78 NBM Wierzba 30.9605 12.0395 43 0.67 NBM Wygryńska 24.55595 -3.55595 21 0.2 Wigry Bryzgiel 56.88256 -10.8826 46 16 Wigry Bryzgiel - ląd 43.20288 -12.2029 31 3.81 Wigry Brzozowa L 10 0.1917 -9.28379 30 2.32 Wigry Brzozowa P = 33.431x y39.28379 23.23839 1.761609 25 0.15 C Cimochowski Grądzik Wigry R² = 0.7215 22.93307 8.066934 31 0.14 N Cimochowski Grądzik Wigry 31.71767 -6.71767 25 0.76 S Cimochowski Grądzik Wigry 60 3.13 Wigry Kamień 41.60497 18.39503 1 34 4.49 Wigry Krowa 44.58461 -10.5846 49 1 36.36101 12.63899 Wigry 0.011.55 Mysia Wigry 100 0.0001 50.60104 13.39896 8.69 Area 64 Wigry Ordów 67.41729 25.58271 93 38.82 Wigry Ostrów 29.91417 -5.91417 24 0.56 Wigry Rośków 31.71767 -3.71767 28 0.76 Wigry Walędziak 57 18 Wigry Wysoki Węgieł 58.18153 -1.18153 The outlier would disturb direct comparisons of species richness Due to possible differences in island areas between the two island complexes we have to use the residuals. A direct t-test on raw data would be erroneous. Upper 2.5% Observed P(t) confidence limit. Frequency Permutation testing 0.08 0.07 0.06 0.05 0.04 0.03 0.02 0.01 0 0 Observed values NBM Wigry 1.778435 -3.55595 -4.27129 -10.8826 2.864436 -12.2029 3.643813 -9.28379 -1.06014 1.761609 -1.04557 8.066934 7.935042 -6.71767 0.929363 18.39503 7.402711 -10.5846 -28.1831 12.63899 15.6314 13.39896 -10.0431 25.58271 0.437301 -5.91417 15.12399 -3.71767 12.0395 -1.18153 t 0.118799 NBM 0.929363 -10.5846 25.58271 1.778435 -3.55595 1.761609 15.6314 -5.91417 -3.71767 -1.04557 13.39896 -9.28379 18.39503 -1.06014 -12.2029 0.34257 Wigry -10.0431 2.864436 3.643813 12.0395 15.12399 8.066934 -4.27129 12.63899 -28.1831 -1.18153 7.402711 -10.8826 0.437301 7.935042 -6.71767 Randomized values NBM Wigry 1.761609 1.778435 7.402711 -3.55595 25.58271 2.864436 -12.2029 -10.5846 -5.91417 18.39503 -10.8826 -9.28379 3.643813 -6.71767 15.6314 13.39896 0.929363 7.935042 -1.18153 0.437301 12.63899 -28.1831 -4.27129 8.066934 -1.04557 -10.0431 12.0395 -3.71767 -1.06014 15.12399 0.766559 NBM -10.5846 15.12399 -12.2029 -4.27129 -1.18153 0.437301 25.58271 -6.71767 15.6314 -9.28379 -1.06014 7.402711 -3.71767 -10.0431 3.643813 0.346264 Wigry -3.55595 1.761609 12.0395 -10.8826 0.929363 1.778435 8.066934 -28.1831 2.864436 12.63899 18.39503 13.39896 7.935042 -1.04557 -5.91417 0.2 0.4 0.6 t-values 0.8 10000 randomizations of observed values gives a null distribution of t-values and associated probability levels with which we compare the observed t. This gives the probability level for our t-test. 1 Bivariate comparisons using ANOVA t 0.11884 F 0.01412 2 2 F t2 t and F tests can both be used for pair wise comparisons. Repeated measures n SSerror ( xij Pi T j x) 2 Species richness of ground living Hymenoptera in a beech forest i 1 j 1 Before Leaf-litter free Plot 34 52 1 39 58 2 1 10 3 52 50 4 45 49 5 6 15 6 33 32 7 12 14 8 28 52 9 1 19 10 35 29 11 7 22 12 33 18 13 7 11 14 9 15 15 10 15 16 3 2 17 7 3 18 T-Test 0.027271 Mean 25.88889 Grand Mean 23 20.11111111 SSEffect 16.69136 df Photo Tim Murray Photo Simon van Noort k 1 k SStreat n (T j x) 2 j 1 Mean 43 48.5 5.5 51 47 10.5 32.5 13 40 10 32 14.5 25.5 9 12 12.5 2.5 5 Mean 43 48.5 5.5 51 47 10.5 32.5 13 40 10 32 14.5 25.5 9 12 12.5 2.5 5 SS Error 74.69136 87.41358 5.191358 30.24691 1.580247 5.191358 22.96914 7.135802 166.0247 74.69136 69.35802 42.52469 215.858 1.580247 0.024691 0.302469 22.96914 47.80247 Sum df SSEffect/ SSError F P(F) 875.5556 17 0.019064 0.324083 0.576609 Advices for using ANOVA: You need a specific hypothesis about your variables. In particular, designs with more than one predicator level (multifactorial designs) have to be stated clearly. ANOVA is a hypothesis testing method. Pattern seeking will in many cases lead to erroneous results. Predicator variables should really measure different things, they should not correlate too highly with each other The general assumptions of the GLM should be fulfilled. In particular predicators should be additive. The distribution of errors should be normal. It is often better to use log-transformed values In monofactorial designs where only one predicator variable is tested it is often preferable to use the non-parametric alternatives to ANOVA, the Kruskal Wallis test. The latter test does not rely on the GLM assumptions but is nearly as powerful as the classical ANOVA. Another non-parametric alternative for multifactorial designs is to use ranked dependent variables. You loose information but become less dependent on the GLM assumptions. ANOVA as the simplest multivariate technique is quite robust against violations of its assumptions.