light
advertisement

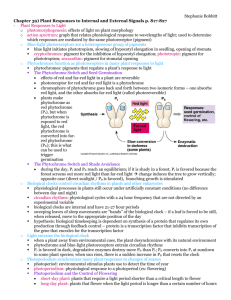
Plant-environment interaction 1: How do plants respond to light? Lecture 1: The phytochrome pathways • Photomorphogenesis -- The light-induced developmental and morphological changes. So light not only serves as energy source for photosynthesis but also as a signal for growth and development. How does plants respond to light signal? Light dark Monocot Light dark dicot 2. Molecular changes during photomorphogenesis From signal (light) to responses at the molecular level Light-induced gene expression—the “greening” process as a model. Etiolated plants---exposed to light---genes that are activated (related to the greening process---building the chloroplasts for photosynthesis). 3. Plants have different receptors to detect various spectra of light Red light receptors and blue light receptors are best defined. The receptors are the beginning step in light signaling pathways. 4. The phytochrome pathway 1) The function of red light in development: seed germination, greening (chloroplast development), regulation of internode elongation, flowering time, bud dormancy etc… 2) The red light receptors—phytochrome family—have two reversible light absorbing forms---Pr for red light-absorbing form and Pfr for far red absorbing form. The Pfr is the biologically active form. 666 Pr Pfr 730nm Red/far red switch in seed germination 3) In arabidopsis there are at least 5 different isoforms of phytochrome PhyA,B,C,D,E. Each of them is a peptide of about 120 kDa. The native form is a dimer of about 250 kDa with chromophore group called phytochromobilin. PhyAE function differently but with some overlap. PhyA and PhyB are better studied Upon red/far red switch, the protein and the chromophore both change conformations 4) The phytochrome domains The chromophore-binding domain and the signaling domain: PEST is for photo-degradation 5) The mechanism of signaling: from phytochrome to gene expression in the nucleus a) Where is phytochrome? From cytosol to nucleus! Much phyA/B protein Are found in the cytosol in the dark but light induce nuclear localization of the Phy proteins, suggesting that downstream events may occur in the nucleus But things are more complicated—studies show that both cytosolic and nuclear pathways exist b) The pathways: i) PhyB directly interacts with the transcriptional factor The yeast two hybrid system: A powerful tool to find the partner proteins—as most proteins function by interacting with other proteins, finding partner proteins is critical for identifying functions of a protein in the cell. Your protein of interest is used as a “bait” to fish out the “prey” in a cDNA library. A partner protein is identified for PhyB called PIF3 (PhyBInteracting Factor #3). Red light causes PhyB to move into nucleus and interact with PIF3, a transcription factor that activates the genes encoding other transcription factors for activation of photosynthetic genes. ii) The phyA serves as a protein kinase and phosphorylates substrates in the cytoplasm The bacterial phytochrome is a histidine kinase: iii) The G protein pathway? phyA mutant rescue by microinjection of G protein pathway components (GTP) and calcium-CaM, and cGMP. iv) What is going on in the nucleus? Protein degradation is a key regulatory process. Identification of a E3 ligase (called COP1) as a negative regulator in the light signaling pathway—mutant causes constitutive photomorphogenesis---the gene encodes a ubiquitin pathway ligase for degrading transcription factors required for light-induced genes expression. v) Other photoreceptors modulate phytochrome pathway