Dr Joanne Chory of The Salk Institute, Howard Hughes Medical
advertisement

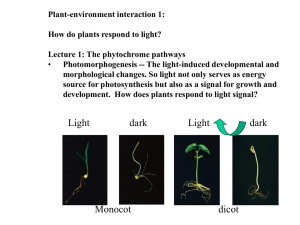
How do light signals control nuclear genes for leaf & plastid development? Can divide into 3 basic steps (or parts): 1. Receiving the signal (photoreceptors) 2. Transmitting (and amplifying?) the signal to the nucleus 3. Activating (de-repressing?) or repressing transcription of genes associated with “greening” or “de-etiolation” Transcriptional control of the pea rbcS3 gene by light:: The molecular approach • Comparative analysis of 5’-upstream sequences of the rbcS gene family (pea) identified light-responsive elements (LRE) Enhancer ______________________________________ III* II* I II III -330 VI -50 V +1 N-H. Chua • Several putative trans-acting factors for this promoter were identified based on their in vitro ability to bind to specific elements - GT1, AF2 & AF3 binds to, or near, boxes II and/or III (and II* and/or III*) - AF1 binds box VI • Present in both light and dark, however. • Some maybe regulated by phosphorylation-dephosphorylation - Binding of AF3 to DNA is promoted by phosphorylation - Kinase may be a casein kinase 2 (CK2) Postive and negative factors from a genetic approach in Arabidopsis Another long hypocotyl mutant, Hy5, lacks a bZIP factor that promotes transcription from a number of genes with LREs - Hy5 also responds to the blue (Cry) and red light (Phy) photoreceptors bZIP proteins have basic (+) DNAbinding domain and a leucine dimerization zipper COP/DET genes: Pleiotropic repressors of leaf and plastid development • Pleiotropy- one gene affects many processes • J. Chory identified a mutant DET1 that de-etiolates even in darkness – cotyledons are not green, but they do expand – plastids develop into chloroplast-like organelles – Many greening genes (rbcS and cab) are on Det1 light Det1 dark WT dark COP/DET/FUS genes: Pleiotropic repressors of leaf and plastid development COP (constitutive photomorphogenesis) and FUS mutants similar to DET1 10 COP genes: - COP1, key repressor of photomorph. - 8 are subunits of a large complex, COP9 signalosome, - COP1 and COP9 complex also found in animal cells COP1 - COP1 is a ubiquitin ligase, which triggers degradation of transcription factors (HY5) by the proteasome, also interacts with COP9 and DET1 Yi and Deng, 2005, TCB 15:618 PhyA, PhyB, Cry1, Cry2 inhibit COP1-mediated degradation of transcription factors that activate photomorph. genes Cry proteins are activated by BL, phosphorylated, and then bind to COP1 COP1 also moves to cytoplasm in light. How do PhyA and PhyB inhibit COP1?? Phytochrome Interacting Factors (PIFs) Another Model for Phytochrome Action. Enamul Huq, UT Peter Quail, USDA PhyB sees Red and PhyA, Far-red D Rc Arabidopsis thaliana mutants FRc WT phyA phyB Tepperman et al., 2004 Two-Hybrid Screening Strategy to get Phy Interacting Factors (PIFs) phy PIFs AD BD GAL1 UAS HIS3/lacZ PIF3 (bHLH protein): Required for Full PhyBMediated Greening Response Pho4 (bHLH) bound to DNA Monte et al (2004) PNAS CCA1 & LHY have G-box Martinez-Garcia et al (2000) Science 288: 859 At least some PhyA and PhyB Translocate into Nucleus in Light phyB phyA Nagy and Schaefer (2000) EMBO J. 19: 157-163. GFP-tagged Phy proteins Postulated Direct Targeting of Light Signal to Promoter-Bound PIF 3 PrB R PIF3 PIF3 G-box TA TA FR PfrB PfrB FR PrB NUCLEUS PfrB PIF3 PIF3 G-box CYTOPLASM PIC TA TA Does PIF3 heterodimerize? A 2-Hybrid Screen with PIF3 as Bait PIF3 BD Y AD HIS3/lacZ GAL1 UAS ** PIF1 and PIF4 are new bHLH proteins What is/are the function of the heterodimer(s)? SUMMARY 1. Interaction of DNA-bound PIF3 with Pfr form of PhyB may provide for direct regulation of gene expression in response to light. 2. PIF3 helps controls greening process, and interacts with other PIFs. 3. Phy signaling involves direct interaction with transcription factors (PIFs).