3723 Exp 10b-Kinetics2
advertisement

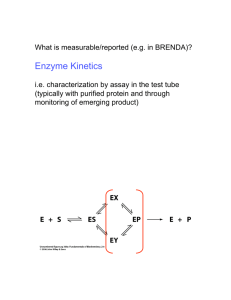
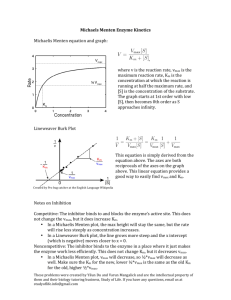
10b Kinetics 3723 F02 1 Biochemistry 3723 Lecture 10b Kinetics Part 2 October 23, 2002 I. Different methods of plotting vo vs. S data to determine kinetic constants A. Michaelis-Menten = non linear regression-Vmax Vo Vmax/2 Km [S] 1. vo = Vmax•[S]/(Km + [S]) 2. Pre-computers: it was hard to determine fundamental kinetic constants from this type of plot (need Vmax to determine Km and Vmax only truly reached at infinite [S]). With computer analysis, non-linear regression becomes most accurate plot to use. 3. Led to reworked rate equations that give linear representation of data. We look at two of them. B. Lineweaver Burk--first developed 1. 1 K 1 1 m v o Vmax [S] Vmax 2. x intercept = -1/Km; y intercept = 1/Vmax; slope = Km/Vmax. C. Eadie Hofstee Vmax v Slop e = -Km Vmax/K m v/S 10b Kinetics 3723 F02 2 1. v o K m vo Vmax [S] 2. dependent variable (v) in both x and y axis. D. [Substrate]--what range? How many different data points? 1. Practical limits: sensitivity at low [S], solubility at high [S] 2. Range? ≈ from Km/2 to 10 Km, spaced more closely at low S (more data points) , with at least one point approaching Vmax. 3. How many replicates? Want standard deviation <10% of mean. 4. How many points? 5 to about 12 [S] depending on the quality of the data. 5. If more than one substrate, will need many more points: minimum: 5 x 5 = 25!! II. Inhibition A. Mechanism and diagnostics--Lineweaver-Burk and Eadie Hofstee plots useful for diagnostics of TYPE of inhibition B. Compare rates of uninhibited reaction to reactions w/ various [inhibitor] 1. Competitive inhibition: Inhibitor binds E only. Can easily imagine substrate analog as competitive inhibitor. a Change Km(app), but not Vma b. Eadie Hofstee and Lineweaver-Burk: y intercept unchanged. Slope and x intercept change. 2. Non-competitive/Mixed: Inhibitor binds E or E•S a. If Ki = Kii, called non-competitive (uncommon) i. Km(app) unchanged; Vmax changed ii L-B; Lines converge on x axis: E-H: Parallel lines 10b Kinetics 3723 F02 3 b. “Mixed” inhibition: non-competitive in which Ki and Kii differ (binding of I to E differs from binding of I to E•S). Common for inhibitors that aren’t substrate analogs and bind away from active site i. both Km(app) and Vmax change ii. L-B intersect not on axis: E-H; non parallel, non intersecting lines iii. Must be careful that this is real and not simply poor data for competitive inhibition. 4. Uncompetitive: Inhibitor affects E•S only. Important for multisubstrate enzymes a. Change apparent Km and Vmax b. L-B; Parallel lines: E. H; Converge at Km/Vmax (x intercept) III. Practical Considerations. A. For today’s exercise: 1. You chose [S] within guidelines given (0.10 to 5 mM NPP) 2. Use [enzyme] determined last lab to be valid for kinetics 3. Assay 1.0 ml (not 0.5 ml); Assay at t = 0 and t = 10 only a. Reasoning: if linear for 20 min., assaying for 10 min. gives added margin of safety. b. Use larger aliquot to give more color--especially for inhibited reactions this will give us better accuracy. 4. Accuracy of pipetting and timing critical to quality of data. B. Data Work-up is mostly by computer. Do at your convenience. Computer program on computers in lab (folder is titled EnzymeKinetics v1.5). Instructions in Appendix V 1. Converts ∆A405/min to ∆product/min for you. But you must know how to do it for an exam! 2. Review: 10b Kinetics 3723 F02 4 A E C l; E 18.8 liter mmol cm mmol A(min 1 ) liter liter min 18.8 1 cm mmol cm mmol 1 liter 10 3 mol 30 ml 5 ml rxn liter • min 1000 ml mmol 5 ml rxn (undiluted) 0.05 ml enzyme moles / min ml enzyme Note: 30 ml/5 ml rxn mix takes into account dilution by NaOH. C. Inhibition constant. 1. To determine inhibition type and constants, need to repeat above analysis using several inhibitor concentrations. We use four (0, 0.25, 0.50. 1.00 mM PO4). 2. Determine Ki: Can do mathematically from rate equation 3. Computer does it graphically: From Eadie Hofstee equation a. Plot 1/(variable intercept) vs. [Inhibitor] Dixon Plot Example for competitive C / min b. Computer values: if numbers make no sense, it’s because they aren’t real. Sample Ki Kii 0.589 -139 LB 0.366 -8.9 EH 0.653 +52.0 HW 0.369 -10.4 JL 0.545 +2140 DL 0.654 +204 NLR Note that the values calculated by the computer for Ki range from 0.37 – 0.67, while the values calculated for Kii range from -140 to +2100, which of course makes no sense (for example Kii cannot be negative). The type of inhibition exhibited by the enzyme for which this data was generated has only one Ki . It is up to the investigator to evaluate whether the data make any sense. Again, these values are controlled by the quality of the original data. The computer will always calculate a Ki and a Kii (even for competitive or uncompetitive inhibition which don't have a Kii). 10b Kinetics 3723 F02 5 IV. Look at Enzyme Kinetics Program--FTP/EnzymeKinetics--In Lecture will walk you through a sample of the calculations. V. Problem--You have done a several series of enzyme assays using different concentrations of substrate and enzyme inhibitor, with the results shown below: a. What kind of plot is shown below? Solution: This is a 1/v vs. 1/S plot, which is a Lineweaver-Burk plot b. How can you determine Km and Vmax from this plot (for the uninhibited reaction) Solution: 1/ Vmax is the y intercept for the line marked no inhibitor. -1/Km is the x intercept. You can solve for Vmax and Km mathematically from these values. c. What kind of inhibition does this plot represent? Name it and draw (using E, S, I, and P) the reaction scheme. Solution: The plot shows a changing Vmax but constant Km. Therefore it is a noncompetitive inhibition (rare, except maybe for in exam questions). d. Show how you can graphically determine the KI for this reaction? Solution: To determine the Ki (Ki and Kii will be identical in this case), plot 1/y intercept (apparent Vmax in this case) on the y axis vs. the inhibitor concentration on the x axis. Extend the line formed by those points to cross the x axis. The x intercept is –Ki.